Cpgplot
- CPGPLOT(+)
CPGPLOT(+)
FUNCTION
CpGPlot plots the frequency of occurence of CpG di-nucleotides and C and G percentage relative to their position in a sequence by the method described by Gardiner-Garden (1987)
DESCRIPTION
CpGPlot plots the observed/expected frequency of CpG di-nucleotides and the percentage of Cs and Gs within a window that steps along the sequence at a specified shift. The method is described in Gardiner-Garden (1987); J.Mol.Biol. 196:261-282.
AUTHOR
This program was written by Rodrigo Lopez S. (E-mail: rodrigol@biotek.uio.no; Post: Biotechnology Centre of Oslo, PO Box 1125 Blindern, N-0317 Oslo 3, Norway).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a session with CpGPlot
% cpgplot -cgline
CpGPlot of what nucleotide sequence ? GenEMBL:Hsh4bhis
Start (* 1 *) ?
End (* 814 *) ?
What window size (* 100 *) ?
What shift increment (* 1 *) ?
What should I call the output file (* hsh4bhis.islands *) ?
The minimum density for a one-page plot is 707.8 bases/100 platen units.
What density do you want (* 707.8 *) ?
%
OUTPUT
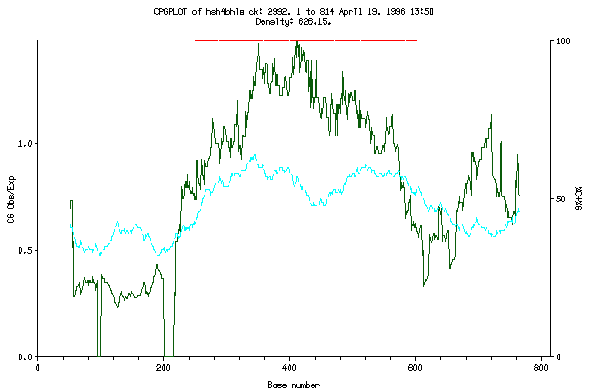
This is the plot from the example session
RESTRICTIONS
Not known
ALGORITHM
The algorithm used is described by Gardiner-Garden (1987); J. Mol. Biol. 196:261-282). The method is based on the calculation of a running average in a window that steps along the sequence at a specified shift. The observed/expected CpG ratio and the percentage of C's and G's is calculated within his window to produce the two numerical arrays plotted by the program.
CONSIDERATIONS
The length of the sequence being analysed must be taken into consideration when interpreting the result of the plot. Increasing the window size and shift will result in smoothing of the data with possible loss of CpG detection. On the other hand, failing to do so may result in plots that are very difficult to interpret.
SUGGESTIONS
Use the default setting for sequences approximately 4000 bases long. Longer sequences may require larger shift and/or window sizes.
GRAPHICS
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
CTRL-C
If you need to stop this program,
use
All parameters for this program may be put on the command line.
Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes.
In the summary below,
the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter.
Square brackets ([ and ])
enclose qualifiers or parameter values that are optional.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
The files described below supply auxiliary data to this program.
The program automatically reads them from a public data directory unless you either 1)
have a data file with exactly the same name in your current working directory;
or 2)
name a file on the command line with an expression like -DATa1=myfile.dat.
For more information see Chapter 4,
Using Data Files in the User's Guide.
The parameters and switches listed below can be set from the command line.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
plots a line indicating CpG righ areas
sets the observed/expected CpG ratio used for island detection.
sets the percent GC used for island detection.
sets the minimum length used for island detection.
plots a line at the observed/expected ratio threshold.
plots a line at the GC percent threshold.
suppresses the %CG line.
specifies an alternative plot title.
suppresses the plot title.
If you are studying a sequence with known features,
this program marks the plot with small boxes showing the positions of these features.
The presence of a file in your directory with the same name as your sequence and the file name extension .mrk causes the program to mark each range specified in the file.
You can provide a marking file on the command line with an expression like -MARk=
hsh4bhis.mrk.
The file gamma.mrk contains information about the format of marking files.
The figure for the example session shows marked regions.
If you are studying a sequence with known features,
this program marks the plot with small boxes showing the positions of these features.
The presence of a file in your directory with the same name as your sequence and the file name extension .mrk causes the program to mark each range specified in the file.
The file gamma.mrk contains information about the format of marking files.
These options apply to all GCG graphics programs.
These and many others are described in detail in Chapter 5,
Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure
program instead of drawing the plot on your plotter.
draws all text characters on the plot using Font 3 (see Appendix I)
.
draws the entire plot with the pen in stall 1.
These options let you expand or reduce the plot (zoom),
move it in either direction (pan),
or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0)
to 1.2 (zoom in).
You can expand the axes independently with -XSCAle and -YSCAle.
Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees.
Usually,
plots are displayed with the horizontal axis longer than the vertical (landscape).
Note that plots are reduced or enlarged,
depending on the platen size,
to fill the page.
Gardiner-Garden,
M (1987).
J.Mol.Biol.
196 261-282.
Printed: April 22,
1996 15:52 (1162)
COMMAND-LINE SUMMARY
Minimum syntax: % cpgplot [-INfile=]GenEmbl:Hsh4bhis -Default
Prompted Parameters:
-BEGin=1 -END=814 the range of interest
-WINdow=100 the window length
-SHIFT=1 the window shift
Local Data Files: None
Optional Parameters:
-CGLine plots a line indicating CpG rich areas
-MINOBSexp=0.6 Obs/Exp threshold for island detection
-MINPC=0.5 percent GC threshold for island detection
-MINlen=200 minimum length for island detection
-SHOWOBSexpline plots a line indicating Obs/Exp threshold
-SHOWPCline plots a line indicating percent GC threshold
-NOPERcent suppresses the percentage CG line
-TITLEText="A Title" alternative plot title
-NOTITle suppresses the plot title
Most EGCG graphics programs accept these and other switches. See the Using
Graphics chapter of the EGCG USERS GUIDE for descriptions.
-DENSity=150.0 plot density in bases per 100 platen units
-LEFTMARgin=10.0 sets the left plot margin position
-RIGHTMARgin=140.0 sets the right plot margin position
-BOTTOMMARgin=10.0 sets the bottom plot margin position
-TOPMARgin=90.0 sets the top plot margin position
-BORDer puts a line border around the plot
-NOBORDer suppresses a line border
-PAGENUMber forces page numbering
-NOPAGENUMber suppresses page numbering
-TITletext="text" overrides the default plot title
-NOTITletext suppresses the plot title
-SUBTITletext="text" overrides the default plot subtitle
-NOSUBTITletext suppresses the plot subtitle
-CHEIGHT=1.5 default plot character height
-LINESTyle1=1 plot line style 1 (set for each line)
-LINEPERiod1=1 plot line period 1 (set for each line)
-LINECOLor1=0 plot line colour 1 (set for each line)
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
LOCAL DATA FILES
OPTIONAL PARAMETERS
-CGline
-MINOBSexp=0.6
-MINOBSexp=50
-MINlen=200
-SHOWOBSexpline
-SHOWPCline
-NOPERcent
-TITLEText="A Title"
-NOTITle
-MARk=hsh4bhis.mrk
-FIGure=programname.figure
-FONT=3
-COLor=1
-SCAle=1.2
-XPAN=30.0
-YPAN=30.0
-PORtrait
REFERENCES