Patternplot
PATTERNPLOT(+)
FUNCTION
PatternPlot produces a graphical representation of the results of GCG's FindPatterns program.
DESCRIPTION
PatternPlot uses a Findpatterns output file, whose extension should be ".find". It displays the location of the nucleotide or peptide patterns found within one (or more) sequences. For each sequence and for each pattern a line is drawn ( labelled relatively to the length of the sequence ). For each occurence of the pattern, a blue rectangle is printed above the sequence line (or a red one beneath in case of reverse strand). The position of a rectangle depends on its position in the sequence, and its length depends on the ratio of the pattern length to the sequence length. Mismatches are displayed in green (instead of red or blue).
AUTHOR
This program was written by Philippe Dessen (E-mail: dessen@infobiogen.fr) and colleagues at the French EMBnet node (Post: INFOBIOGEN, 7 rue Guy Moquet - BP8, 94801 Villejuif CEDEX, France).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a session using Patternplot to display the results of the file findpatterns.find:
% patternplot PATTERNPLOT of what file ? test.find PostScript instructions for a LASERWRITER are now being sent to plot.ps %
RELATED PROGRAMS
PatternPlot is a graphic extension of FindPatterns, and therefore requires no special options. All the choices are to be made when using Findpatterns, as the choice of the sequences or patterns and the possibility of allowing mismatching.
RESTRICTIONS
This version of PatternPlot cannot support files containing patterns longer than approximately 200 characters. Concerning longer patterns, PatternPlot does not seem to be very relevant because in that case the information of interest is the location, which clearly appears in the output of FindPatterns.
INPUT FILES
As an extension of FindPatterns, PatternPlot uses files with a ".find" extension.
OUTPUT
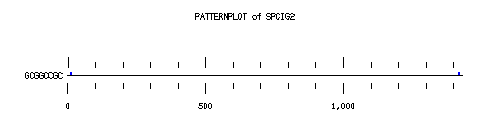
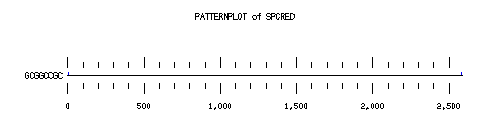
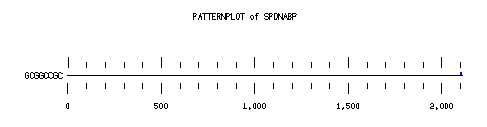
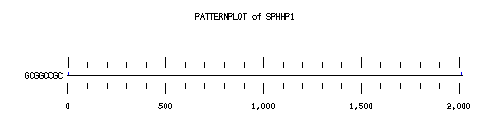
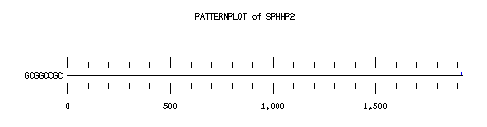
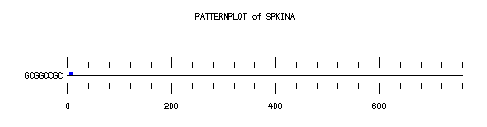

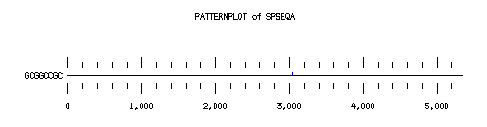
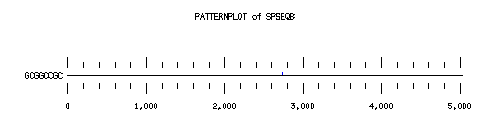
These are the plots from the example session
LOCAL DATA FILES
None.
OPTIONAL PARAMETERS
None.