Phylip2tree
PHYLIP2TREE
FUNCTION
Phylip2Tree displays trees computed with one of the PHYLIP-programs in GCG style.
DESCRIPTION
Phylip2Tree draws trees computed by the Phylip programs in GCG style, and displays multiple trees on different pages. This program has been written by Karin Harder from DKFZ, Heidelberg, Germany. The program was further modified for EGCG by Peter Rice (E-mail: pmr@sanger.ac.uk Post: Informatics Division, The Sanger Centre, Hinxton Hall, Cambridge, CB10 1RQ, UK).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a sample session with Phylip2Tree mono % phylip2tree %
OUTPUT
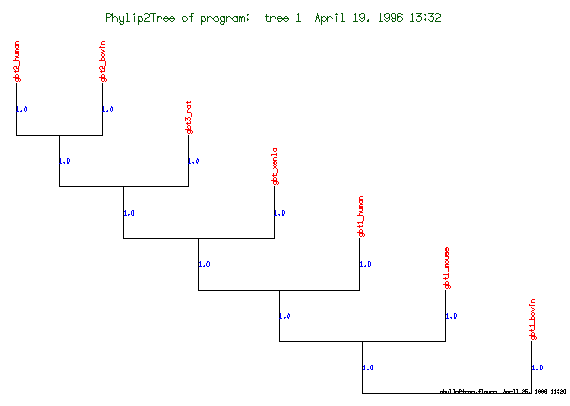
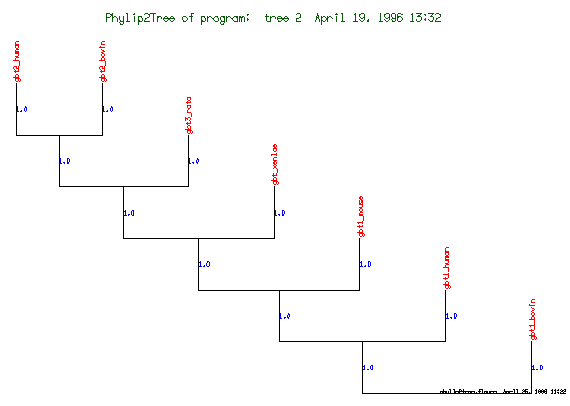
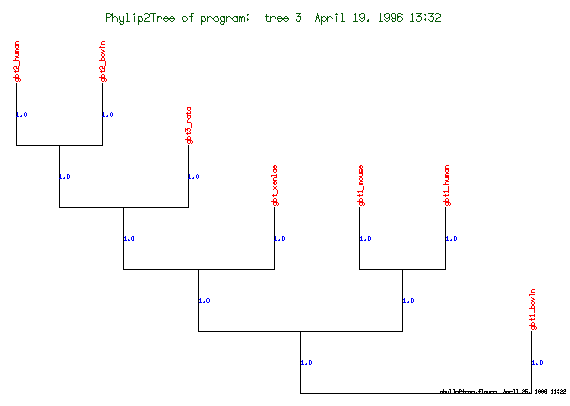
These are the plots from the example session
INPUT FILE
The input file for phylip2tree is an output file from a Phylip program.
COMMAND-LINE SUMMARY
All parameters for this program may be put on the command line. Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes. In the summary below, the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose qualifiers or parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
Mimimum Syntax: % phylip2tree [-INfile=]treefile -Default Required Parameters: -PROGName=DnaPars name of program which computed treefile Optional Parameters: none
Printed: April 22, 1996 15:54 (1162)