Plotalign
- PLOTALIGN
PLOTALIGN
FUNCTION
PlotAlign takes a GCG format sequence alignment, and plots the mean and range of values for any amino acid parameter you supply. The "panel file" contains a list of parameters to be plotted. The main database of parameters is taken from Nakai et al. (1988), and the default panel file uses selected parameters from the 13 discrete clusters in that paper. This program is experimental. Any suggestions would be most welcome.
DESCRIPTION
PlotAlign is an improved version of the EGCG program PrettyPlot. PrettyPlot was originally written to produce publication-quality sequence alignment output. PlotAlign in addition plots any selected amino acid residue parameters (mean, minimum and maximum values, or mean and standard deviation) below the sequence alignment.
PlotAlign plots sequences and the calculated amino acid parameters with their columns aligned. This utility is used after a number of sequences have had gaps added to make them all align. PlotAlign s output allows you to look at relationships among several sequences and to identify the conserved properties in common at each residue position. You should use a file of sequence names to define the sequences you want PlotAlign to display (see Appendix VI). Although a specification such as "*.pep" is also accepted you would not be able to use the various weighting and naming options described below.
ACKNOWLEDGEMENTS
The original suggestions for the PlotAlign program were from Franc Pattus and Peter Sibbald at EMBL.
AUTHOR
This program was written by Peter Rice (E-mail: pmr@sanger.ac.uk Post: Informatics Division, The Sanger Centre, Hinxton Hall, Cambridge, CB10 1RQ, UK).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
By repeatedly using the program Gap with the command line option -OUT, gaps were added to a group of picorna virus capsid proteins in the antigenic region to make them align with each other and with a growing consensus sequence.
This procedure can be replaced by any multiple sequence alignment procedure such as Mali (Martin Vingron), ClustalW (Des Higgins), MSE (Will Gilbert) or the new PileUp program in GCG version 7. The resulting sequence files must be converted to GCG format before use in PrettyPlot, typically by extracting the individual sequences with an editor, creating individual sequence files beginning with ".." on the first line, and converting to GCG format with the Reformat program.
% plotalign
PLOTALIGN uses protein sequences
PLOTALIGN of what sequence(s) ? @pretty.list
Fa10.Ugly, len: 349
Fa12.Ugly, len: 349
///////////////////
R14.Ugly, len: 349
R2.Ugly, len: 349
Start (* 1 *) ?
End (* 349 *) ?
Find consensus to what minimum plurality (* 2.0 *) ?
%
OUTPUT
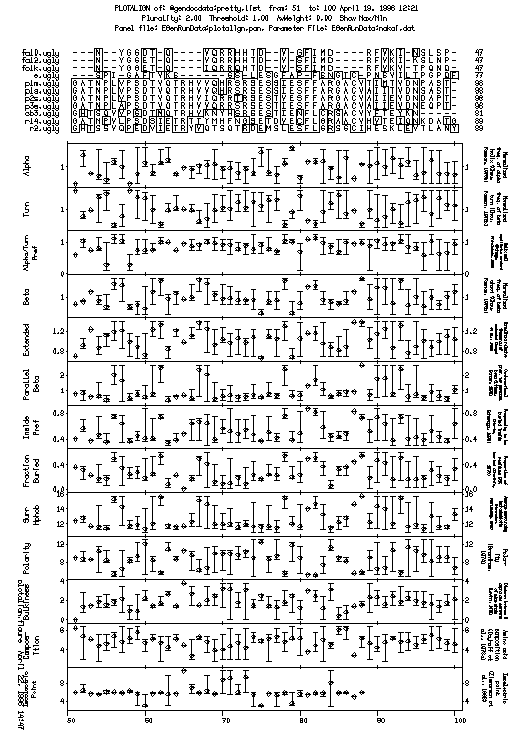
This is the plot from the example session
RELATED PROGRAMS
If you run Gap with the command line options for sequence output, it will write sequence files with the sequences expanded by the addition of gaps. Only two sequences can be aligned at once. LineUp is an editor that allows you to edit multiple sequence alignments.
Mali is Martin Vingron's Multiple Alignment program. ClustalW is Des Higgins' multiple alignment program. MSE is Will Gilbert's multiple alignment editor. All these programs are available from the EBI Network File Server (NETSERV@ebi.ac.uk).
PrettyPlot and PrettyBox will plot boxed sequence alignments as graphics output in a similar way to Pretty which only produces text output files.
RESTRICTIONS
PlotAlign displays sequences which have already been aligned. You can use up to 200 sequences with up to 20,000 symbols in each sequence. This restriction is easily increased on most systems.
CALCULATING A CONSENSUS
PlotAlign calculates a consensus for the column using a symbol comparison table called PrettyPep.Cmp. The consensus is found by finding the symbol in the column for which its comparison to all of the symbols in the column (including itself) yields the greatest number of votes. A vote is cast for each symbol comparison that is over some set threshold value. The votes can be either 1.0 or some "vote weight" assigned to the sequence from which the vote comes.
If there is no coalition of votes that is larger than all of the other coalitions or if the largest coalition is below the minimum plurality, then there is a choice of consensus for the column. By default, no consensus is then displayed. The -NOCOLLision option makes PlotAlign box all possible consensus matches, and choose the first one found for use in the consensus sequence.
The weights for each sequence, the threshold, and the minimum plurality are all real numbers.
If you use -CONsensus, PlotAlign will add a line to your alignments with a "consensus" sequence. The consensus is the symbol that had the largest number of votes (vote weights) in the column. The consensus is included in both the text and graphics versions of the output.
-THReshold=1.0
determines the symbol comparison value below which a symbol may not vote for a coalition. Please note that in the default comparison table an exact match between two amino acid residues scores 1.5, and that some other pairs (D and E, W and Y, L and F for example) are also, by default, considered to match. You should specify -THReshold= 1.5 to force exact matches only.
-PLUrality=2.0
defines the number of votes (vote weights) below which there will be no consensus. By default, each sequence has a vote of 1.0 in creating the consensus.
Vote Weight
If several of your sequences are very similar, you may not want their votes to dominate the consensus for the column. If your input file specification to PlotAlign is a file of file names, you can assign each sequence a vote weight by adding a number to the line after the sequence name. The vote weight is the vote that each row casts for the consensus. Here is the file of file names used to run the example above. Note how each kind of sequence is assigned a vote weight so that their combined impact on the election is never more than one vote.
INPUT FILE
Multiple sequence alignments are best represented with files of sequence names. For PlotAlign these files may include a vote weight as a column of numbers. Here is the input file (pretty.list) from the example session:
A multiple sequence alignment represented as a list file for input to the programs PRETTY, PROFILEMAKE and LINEUP. 7/30/94 .. GenDocData:fa10.ugly wgt: 0.5 GenDocData:fa12.ugly wgt: 0.5 GenDocData:fo1k.ugly wgt: 1.0 GenDocData:e.ugly wgt: 1.0 GenDocData:p1m.ugly wgt: 0.25 GenDocData:p1s.ugly wgt: 0.25 GenDocData:p2s.ugly wgt: 0.25 GenDocData:p3s.ugly wgt: 0.25 GenDocData:cb3.ugly wgt: 1.0 GenDocData:r14.ugly wgt: 0.5 GenDocData:r2.ugly wgt: 0.5
COMMAND-LINE SUMMARY
All parameters for this program may be put on the command line. Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes. In the summary below, the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose qualifiers or parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
Minimum syntax: % plotalign [-INfile=]@Pretty.Fil -Default Prompted Parameters: -BEGin=1 -END=349 range of interest Local Data Files: [-VOTES=]prettypep.cmp comparison table for consensus [-PANELs=]plotalign.pan list of parameter codes and descriptions [-PARAMeters=]nakai.dat list of parameter values Optional Parameters: -JOIN plot line between mean value points -TRIM=0 ignore 1 or more high and low values -SD plot mean and standard deviation -SHOWSEQ=100 maximum number of sequences to be displayed -THReshold=1.0 sets min value for symbol to vote in consensus -PLUrality=2.0 defines the minimum number of votes for a consensus -NOCOLlisions box alternative consensus residues -LINESize=50 sets the number of residues per page -DENSity=50 same as -LINESize -NONUMber no sequence numbering on right of plot -NONAME no sequence name on left of plot -NOTITLE no title at top of plot -DIFferences[="-"] only shows positions disagreeing with the consensus -NOCOLLisions allows more than one alternative consensus residue
LOCAL DATA FILES
The files described below supply auxiliary data to this program. The program automatically reads them from a public data directory unless you either 1) have a data file with exactly the same name in your current working directory; or 2) name a file on the command line with an expression like -DATa1=myfile.dat. For more information see Chapter 4, Using Data Files in the User's Guide.
PlotAlign calculates a consensus for each column using a symbol comparison table (Appendix II) . You can provide your own table called PrettyPep.Cmp. You can define some other table with the command line specification -VOTes=FileName.
OPTIONAL PARAMETERS
The parameters and switches listed below can be set from the command line. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
-LINESize=50
specifies the number of sequence symbols to display on each page. Typically a linesize of 50 is used, but any value from 40 to 80 is reasonable on most output devices.
-CONsensus
causes PlotAlign to show a consensus sequence for the set of sequences you are displaying. Read how PlotAlign finds the consensus above.
-THReshold=1.0
determines the symbol comparison value below which a symbol may not vote for a coalition. See the topic called CALCULATING A CONSENSUS.
-PLUrality=2.0
defines the number of votes (vote weights) below which there will be no consensus. See the topic called CALCULATING A CONSENSUS.
-NONUMber
removes the sequence numbering from the alignment at the top of the page.
-NONAME
removes the sequence names from the graphics output.
-NOTITLE
removes the title lines from the graphics output.
-PANELs=PlotAlign.Pan
specifies an alternative name for the Panel selection file.
-PARAMeters=Nakai.Dat
specifies an alternative name for the parameter database.
-JOIN
draws a line through the mean values. This is intended for use with the -NORANGE qualifier.
-NORANGE
removes the range bars (minimum and maximum values). The remaining mean values are usually linked with the -JOIN qualifier.
-TRIM=0
removes one or more extreme values (both minimum and maximum) to reduce the impact of "outlier" values and questionable regions of the alignment on the interpretation of the plot.
-SD
plots the standard deviation instead of the minimum and maximum parameter values. This may be more useful for very large numbers of aligned sequences, but experience is limited so far. The values are restricted to the minimum and maximum possible parameter values to keep the plot within the panel width.
-SHOWSEQ=200
large numbers of sequences in an alignment can take up to half of the plot. The number of sequences plotted can be reduced to leave more room for the parameter panels.
-NOCOLLisions
allows more than one alternative consensus to be boxed in the alignment.
-DENSity=50
has the same effect as -LINESize= 50.
Printed: April 22, 1996 15:54 (1162)