Poland
POLAND
FUNCTION
The program Poland simulates transition curves of double-stranded nucleic acids (DNA as well as RNA) . Calculation is based on Poland D. (1974) 'Recursion Relation Generation of Probability Profiles for Specific-Sequence Macromolecules with Long-Range Correlations'.
DESCRIPTION
Poland simulates transition curves of double-stranded nucleic acids (DNA as well as RNA) This program has been written by Gerhard Steger from the University of Duesseldorf, Germany. The adaption to HUSAR/GCG (especially the graphical display) was by Gerd Raether, DKFZ, Heidelberg, Germany. The program was further modified for EGCG by Peter Rice (E-mail: pmr@sanger.ac.uk Post: Informatics Division, The Sanger Centre, Hinxton Hall, Cambridge, CB10 1RQ, UK).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a sample session with Poland
% poland
POLAND uses nucleotide sequence data
POLAND of which sequence ? GenEMBL:ptva
Start (* 1 *) ?
End (* 359 *) ?
Which tables do you want to use?
a) 3DPlot
b) GelPlot
c) MeltPlot
d) TempPlot
Please choose one or more (* ABCD *):
Which thermodynamic parameters do you want to use?
a) Freier et al. (RNA in 1 M NaCl)
b) Poerschke et al. (RNA in 1 M NaCl)
c) Gotoh (DNA in 0.019 M NaCl)
d) Breslauer et al. (DNA in 1 M NaCl)
e) Klump (DNA in 0.1 M NaCl)
Please choose just one (* C *): a
Enter Lowest temperature (* 80.0 *) 100.0
Enter Highest temperature (* 120.0*)
Enter temperature step (* 1.0 *) 2.0
Process set to plot with HP7550 attached to HPGL.OUT
using the HPGL graphic interface.
HPGL instructions for a HP7550 are now being sent to Hpgl.Out.
%
OUTPUT
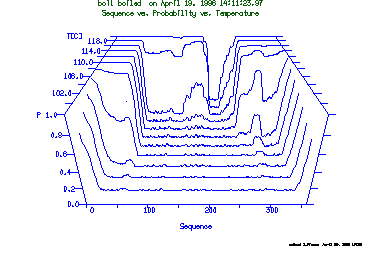
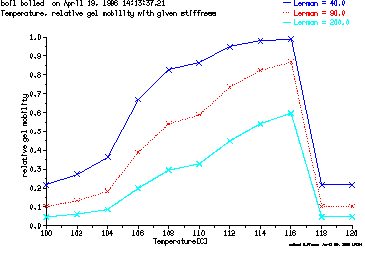
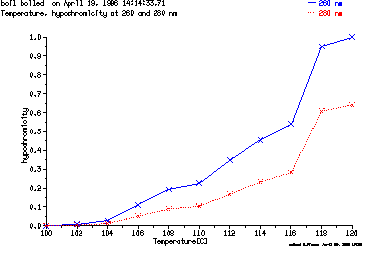
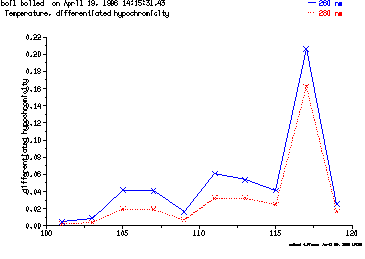

These are the plots from the example session
ALGORITHM
Poland Calculation is based on Poland D. (1974) Biopolymers 13, 1859-1871. 'Recursion Relation Generation of Probability Profiles for Specific-Sequence Macromolecules with Long-Range Correlations' and Fixman and Freire (1977) Biopolymers 16, 2693-2704. 'Theory of DNA melting curves'.
With the original algorithm of Poland (default) computing time is proportional to the sqare of the sequence length.
With -FIXMAN you can change the default algorithm (only in the case of option -LOOP= B) to the modified algorithm of Fixman & Freire. This results in computing time proportional to 10 times the sequence length but works only with loop parameters according to Poland (-LOOP= B).
INPUT FILE
The input file for Poland is a GCG nucleic acid sequence file.
RELATED PROGRAMS
see FOLD: calculates secondary structures of single stranded RNA.
PRIME: GCG 8.x oligonucleotide primer design program.
RESTRICTIONS
The sequence has to be shorter than 1001 nucleotides but longer than 5 nucleotides. In case of a longer sequence she may be resticted by options -BEGin= and/or -END=.
Valid nucleotides are A, G, C, U, and T.
Calculation of asymmetric or bulge loops is not possible.
SUGGESTIONS
Hints for combination of parameters and their values:
DNA
Thermodynamic values according to Gotoh et al. and Klump, both,are ideally suited for calculations. The parameter set of Breslauer et al. does not fit our experiments (?).
Ionic strength dependance
Following values may be used for correction of calculated Tm-values:
Tm2 - Tm1
---------- = f(G:C)*I(G:C) + (1-f(G:C))*I(A:U)
log(c2/c1)
with Tm = transition (midpoint, melting) temperature
c = ionic strength (=concentration of Na ions)
f(G:C) = G:C content
I(X:Y) = dependence of ionic strength of base pair type X:Y
DNA
I(A:T) = 18.3 oC (Owen, R.J., Hill, L.R. & Lapage, S.P. (1969)
Biopolymers 7, 503-516.)
I(G:C) = 11.3 oC (Frank-Kamenetskii, M.D. (1971) Biopolymers
10, 2623-2624.)
RNA
I(A:U) = 20.0 oC (Steger, G., Mueller, H. & Riesner, D. (1980)
I(G:C) = 8.4 oC Biochim. Biophys. Acta 606, 274-284.)
RNA
Thermodynamic values according to Turner et al. are ideally suited for calculation in 1 M ionic strength after correction (see below) of all DeltaS values by 1.021 and all DeltaS(GC) values by 0.961. These corrections are equivalent to a shift in Tm values of A:U stacks by -7 K or -2%, respectively and of G:C/G:C stacks by +7 K or +2%, respectively.
COMMAND-LINE SUMMARY
All parameters for this program may be put on the command line. Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes. In the summary below, the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose qualifiers or parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
Command Syntax: > POLAND [-INfile]=a.seq -Default
Required Parameters:
-BEGin=1 begin of the sequence.
-END=444 end of the sequence
-MENu=abcd sets type of plots/output
a = 3DPlot ( temperature vs. probability vs. position)
b = GelPlot ( relative mobility vs. temperature )
c = MeltPlot ( hypochromicity vs. temperature )
d = TempPlot ( temperature of 50% probab. vs. position )
-THERMo=c sets thermodynamic parameters (only one) according to
a) Freier et al. (RNA in 1 M NaCl)
b) Poerschke et al. (RNA in 1 M NaCl)
c) Gotoh (DNA in 0.019 M NaCl)
d) Breslauer et al. (DNA in 1 M NaCl)
e) Klump (DNA in 0.1 M NaCl)
-MINTem=60. minimum temperature
-MAXTem=80. maximum temperature
-TEMPSTEP=1. temperature step
Optional parameters:
-OUTfile=a.out writes the results to a file
-NOPLOT suppresses the plot
-MISPOS=1,2,3 allow mismatches on defined positions
-SFactor=1.0 correction factor for DeltaS
-SAUfactor=1.0 correction factor for DeltaS(A:U)
-SGCfactor=1.0 correction factor for DeltaS(G:C)
-BETA=1.e-3 dissociation constant
-C0=1.e-6 concentration of single strand
-LOOP=B calculation methods for loops appearing during denaturation:
a) according to Gralla & Crothers
b) according to Poland
c) according to Freier et.al.
-SIGma=1.e-3 correction factor for loop computation
-LR=40,90,200 stiffness of nucleic acid (acc. to Lerman)
-FIXman changes default algorithm (only in case -LOOP=B)
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
LOCAL DATA FILES
The files described below supply auxiliary data to this program. The program automatically reads them from a public data directory unless you either 1) have a data file with exactly the same name in your current working directory; or 2) name a file on the command line with an expression like -DATa1=myfile.dat. For more information see Chapter 4, Using Data Files in the User's Guide.
OPTIONAL PARAMETERS
The parameters and switches listed below can be set from the command line. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
-OUTfile=a.out
writes the results to a file (disabled at present).
-NOPLOT
suppresses the plot.
-MISPOS=1,2,3
allows mismatches at defined positions.
-SFactor=1.0
sets the correction factor for DeltaS.
-SAUfactor=1.0
sets the correction factor for DeltaS(A:U).
-SGCfactor=1.0
sets the correction factor for DeltaS(G:C).
-BETA=1.e-3
sets the dissociation constant.
-C0=1.e-6
(C zero) sets the concentration for a single strand.
-LOOP=B
sets the calculation method for loops appearing during denaturation (A:Gralla and Crothers, B Poland C:Freier et al.).
-SIGma=1.e-3
sets the correction factor for loop computation.
-LP=40,90,200
sets the stiffness (According to Lerman) of nucleic acid.
-FIXman
changes the default algorithm for the Poland calculation (-LOOP= B only).
REFERENCES
Poland D. (1974) "Recursion Relation Generation of Probability Profiles for Specific-Sequence Macromolecules with Long-Range Correlations." Biopolymers 13, 1859-1871.
Fixman and Freire (1977) "Theory of DNA melting curves." Biopolymers 16, 2693-2704.
Owen, R.J., Hill, L.R. & Lapage, S.P. (1969) Biopolymers 7, 503-516.
Frank-Kamenetskii, M.D. (1971) Biopolymers 10, 2623-2624.
Steger, G., Mueller, H. & Riesner, D. (1980) Biochim. Biophys. Acta 606, 274-284.
Printed: April 22, 1996 15:54 (1162)