Polydot
- POLYDOT(+)
POLYDOT(+)
FUNCTION
PolyDot compares two sets of sequences, draws a dotplot for each pair of sequences, and reports all identical matches of a specified length.
DESCRIPTION
PolyDot compares a set of sequences against itself, or against a second set.
The sequences can be nucleotide or protein, aligned or unaligned. The original applications are to compare members of a protein family an to compare the major contigs of a fragment assembly project.
PolyDot uses a word (ktup) comparison method, so that only exact matches of a given minimum length are reported. This makes it more appropriate for closely related sequences.
The output from PolyDot includes all the dotplots on a single page and to the same scale, a list of all identical regions of at least the selected length, and an input file for Segments to report the complete alignment around each detected match.
AUTHOR
This program was written by Peter Rice (E-mail: pmr@sanger.ac.uk Post: Informatics Division, The Sanger Centre, Hinxton Hall, Cambridge, CB10 1RQ, UK).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a session with PolyDot
% polydot
POLYDOT uses any sequences
POLYDOT of what sequence(s) ? @eclac.fil
Reverse (* No *) ?
EGenRunData:eclac.seq len: 7477 wgt: 1.00
EGenRunData:eclaca.seq len: 1832 wgt: 1.00
EGenRunData:eclaci.seq len: 1113 wgt: 1.00
EGenRunData:eclacy.seq len: 1500 wgt: 1.00
EGenRunData:eclacz.seq len: 3078 wgt: 1.00
What word size (* 15 *) ?
What should I call the output file (* eclac.poly *) ?
PostScript instructions for a LASERWRITER are now being sent to gcgplot.ps.
%
OUTPUT
The output from a session with PolyDot is a graphics output file, a list of hits (file extension ".poly"). The command line option -WORDList also generates a set of input files for the GCG program Segments with the file extension ".word".
Part of the output from the example is shown below:
EGenRunData:eclac.seq (7477) vs. EGenRunData:eclac.seq (7477)
1 7477 1 7477 7477
EGenRunData:eclac.seq (7477) vs. EGenRunData:eclaca.seq (1832)
5646 7477 1 1832 1832
EGenRunData:eclac.seq (7477) vs. EGenRunData:eclaci.seq (1113)
49 1161 1 1113 1113
EGenRunData:eclac.seq (7477) vs. EGenRunData:eclacy.seq (1500)
4305 5804 1 1500 1500
EGenRunData:eclac.seq (7477) vs. EGenRunData:eclacz.seq (3078)
1287 4364 1 3078 3078
//////////////////////////////////
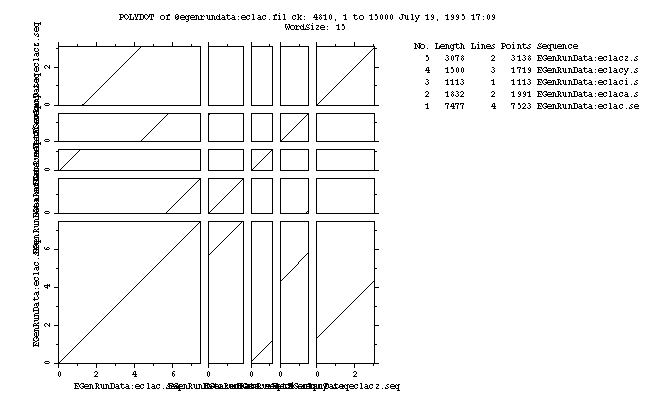
This is the plot from the example session
CONSIDERATIONS
PolyDot can only find a match if there is a minimum number of bases or residues matching perfectly between two sequences. This limits its use to closely related sequences, but allows it to run extremely quickly. Comparisons of all contigs in a cosmid sequencing project (about 40,000 bases) can take only a few seconds to run with a suitable word size.
PolyDot can run with a very small word size to detect weaker similarities, but this drastically increases the number of lines plotted and the run time required.
SUGGESTIONS
A word value of 15 is reasonable for finding direct (tandem) repeats in DNA sequences, or for inverted repeats if one set of sequences is reversed.
The -REVerse option is always worth using in a second run. Remember when comparing contigs for a fragment assembly project that the orientation of the contigs is usually unknown and that you will need a comparison in each direction.
When comparing contigs in a fragment assembly project, the qualifier -MINLENgth is very useful for excluding short fragments which are too small to see on a cosmid scale plot.
INPUT FILE
The input files for PolyDot are nucleotide or protein sequence files.
GRAPHICS
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
CTRL-C
If you need to stop this program,
use
All parameters for this program may be put on the command line.
Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes.
In the summary below,
the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter.
Square brackets ([ and ])
enclose qualifiers or parameter values that are optional.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
None.
The parameters and switches listed below can be set from the command line.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
The second set of sequences can be reversed.
This is very useful when comparing a set of sequences to itself.
sets the name of the main output file.
sets the window size.
A value of 15 is reasonable for DNA sequencing projects,
but a lower value (for example 7)
may be useful for more sensitive but slower comparisons.
sets a minimum length for the plot.
All sequences shorter than this length (small single fragments in an assembly project for example)
will be ignored.
tells PolyDot to write "wordsearch" files for each sequence that has a match above a minimum length.
This length can be more than the word size used for the dot plot.
See the option -MINSCore
below for more detail.
sets the name of each "wordsearch" output file.
The default name uses the sequence name and the extension ".word" which should be good enough for most purposes.
When a large number of lines is found on a plot (usually with a small word size,
or a very large number of simple repeats)
the program can take a very long time to run.
This option reduces the maximum number of lines on any one dotplot to a smaller size,
possibly allowing other plots to show significant matches.
If the plot is truncated,
the bottom left corner (near the start of each sequence)
is drawn.
This limit is to the number of lines drawn on the plot.
The lines can contain any number of points (matching residues).
allows the program to produce the list of hits (and possibly the "wordsearch" output files)
without drawing the actual dot plots.
allows the programs to produce the single page of dot plots without writing the list of all the matches (the ".poly" file).
This can be very useful when testing small word sizes to avoid creating a large output file.
sets the gap between individual dot plots in GCG's "platen units".
The page is "100 platen units high" for a GCG plot.
sets the number of diagonals to be counted together when producing the "wordsearch" output files (with the -WORDList
option on the command line).
This can be set to the number of gaps expected in aligning two contigs or used to reduce the effect of simple repeats.
sets the minimum score used when producing the "wordsearch" output files (with the -WORDList
option on the command line).
The minimum score defaults to the word size,
so any diagonal will be reported.
A higher score forces "wordsearch" hits to have more than the minimum word match.
These options apply to all GCG graphics programs.
These and many others are described in detail in Chapter 5,
Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure
program instead of drawing the plot on your plotter.
draws all text characters on the plot using Font 3 (see Appendix I)
.
draws the entire plot with the pen in stall 1.
These options let you expand or reduce the plot (zoom),
move it in either direction (pan),
or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0)
to 1.2 (zoom in).
You can expand the axes independently with -XSCAle and -YSCAle.
Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees.
Usually,
plots are displayed with the horizontal axis longer than the vertical (landscape).
Note that plots are reduced or enlarged,
depending on the platen size,
to fill the page.
Printed: April 22,
1996 15:55 (1162)
COMMAND-LINE SUMMARY
Minimum syntax: % polydot [-INfile=]genembl:eclac* -Default
Prompted Parameters:
-REVerse Reverses the first set of DNA sequences
-WORDSize=15 Comparison word size (minimum match)
-OUTfile=seqname.poly Output file name
Local Data Files: None
Optional Parameters:
-MINLENgth=1500 Ignores short sequences
-WORDList Write "wordsearch" files for each sequence
-OUTfile2=seqname.word Names of "wordsearch" files
-POINTLIMIT=200000 Maximum number of lines plotted
-NOPLOT Do not produce the dotplot
-NOHITlist Do not produce the list of identity hits
-INTerval=2 Gap between dotplots in platen units
-MAXGAPs=3 Gaps allowed in "wordsearch" matches
-MINSCore=15 Minimum score for "wordsearch" matches
Most EGCG graphics programs accept these and other switches. See the Using
Graphics chapter of the EGCG USERS GUIDE for descriptions.
-DENSity=150.0 plot density in bases per 100 platen units
-LEFTMARgin=10.0 sets the left plot margin position
-RIGHTMARgin=140.0 sets the right plot margin position
-BOTTOMMARgin=10.0 sets the bottom plot margin position
-TOPMARgin=90.0 sets the top plot margin position
-BORDer puts a line border around the plot
-NOBORDer suppresses a line border
-PAGENUMber forces page numbering
-NOPAGENUMber suppresses page numbering
-TITletext="text" overrides the default plot title
-NOTITletext suppresses the plot title
-SUBTITletext="text" overrides the default plot subtitle
-NOSUBTITletext suppresses the plot subtitle
-CHEIGHT=1.5 default plot character height
-LINESTyle1=1 plot line style 1 (set for each line)
-LINEPERiod1=1 plot line period 1 (set for each line)
-LINECOLor1=0 plot line colour 1 (set for each line)
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
LOCAL DATA FILES
OPTIONAL PARAMETERS
-REVerse
-OUTfile=seqname.poly
-WINdow=15
-MINLENgth=0
-WORDList
-OUTfile2=seqname.word
-POINTLIMIT=200000
-NOPLOT
-NOHITlist
-INTerval=1
-MAXGAPs=3
-MINSCore=15
-FIGure=programname.figure
-FONT=3
-COLor=1
-SCAle=1.2
-XPAN=30.0
-YPAN=30.0
-PORtrait