Profileplot
PROFILEPLOT(+)
FUNCTION
ProfilePlot produces a graphical report of the frequency of patterns in a protein or nucleotide sequence.
DESCRIPTION
ProfilePlot plots the "frequency" of patterns (possibly defined using regular expressions) in a sequence (possibly a peptide one). Up to four patterns can be displayed simultaneously.
AUTHOR
This program was written by Philippe Dessen (E-mail: dessen@infobiogen.fr) and colleagues at the French EMBnet node (Post: INFOBIOGEN, 7 rue Guy Moquet - BP8, 94801 Villejuif CEDEX, France).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a sample session with ProfilePlot
% profileplot -lim -win=100 -shi=2
PROFILEPLOT uses any sequence data
PROFILEPLOT of what sequence: ? GenEMBL:hsmyopka
Pattern 1: (AT){1,}
Lower limit (* 0 *):
Upper Limit (* 1 *): 0.2
Pattern 2: T(AA,GA,AG)
Lower limit (* 0 *):
Upper Limit (* 1 *): 0.2
Pattern 3: CG
Lower limit (* 0 *):
Upper Limit (* 1 *): 0.4
Pattern 4: ?
PostScript instructions for a LASERWRITER are now being sent to gcgplot.ps.
%
OUTPUT
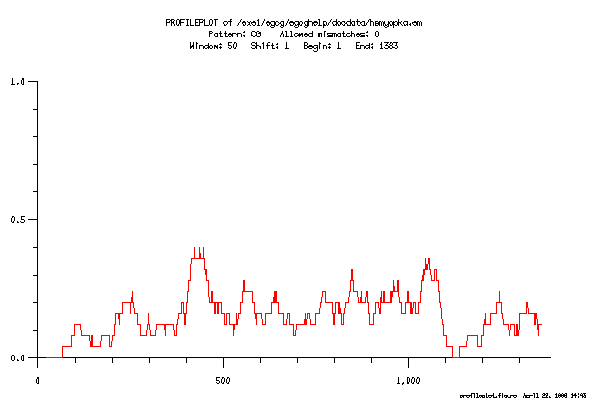
This is the plot from the example session
INPUT FILE
The input file of ProfilePlot is a GCG formatted nucleic acid or peptide sequence.
CALCULATING FREQUENCY
ProfilePlot calculates frequencies over a window. This window can be defined using the qualifiers -WINdow (setting the window size) and -SHIft setting the shift increment used to move the window over the sequence within its range of interest).
The search for patterns matching the regular expresssion in the window begins at the first base of the window. If there is no match, the search restarts on the next base. If several matches are found the shorter is chosen (because several patterns can match a single regular expression). Its length is added to the counter of occurrence then the search restarts at the end of the found pattern. When the end of the window is reached, the counter of occurrence is divided by the window size which gives as a result the frequency.
PLOTTING FREQUENCY
Each pattern has its own graph. The graphic window is divided relatively to their number. If you specified the -LIMit command-line parameter, the limits of the frequency scale will be asked during the pattern specification (as shown in the example), allowing you to have a more precise view for rare patterns.
GRAPHICS
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
CTRL-C
If you need to stop this program,
use
All parameters for this program may be put on the command line.
Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes.
In the summary below,
the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter.
Square brackets ([ and ])
enclose qualifiers or parameter values that are optional.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
The parameters and switches listed below can be set from the command line.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
Specifies the window size.
Specifies the shift increment (used to move the window over the sequence).
Allows the specification of a frequency scale for each pattern.
Allows mismatches during pattern recognition (careful use recommended!).
Forces the program to treat the sequence as a peptide.
COMMAND-LINE SUMMARY
Minimum Syntax: % profileplot [-INfile=]gb_pr:hummyopka -Default
Prompted Parameters:
-BEGin=1 -END=500 the range of interest
-WINdow=50 the window length
-SHIft=1 the window shift
-LIMit allows changing scale
-MISmatch=0 allowed mismatches during pattern recognition
-PROtein treats the sequence as peptide
Local Data Files: None
Optional Parameters: None
OPTIONAL PARAMETERS
-WINdow=50
-SHIft=1
-LIMit
-MISmatch=0
-PROtein=0