RNA Catalysis & Folding
Return to Ongoing Research TopicsRNA Catalysis
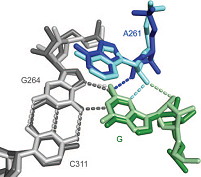 With the discovery nearly thirty years ago that RNA can catalyze reactions with proficiencies that approach those of protein enzymes, the central dogma of biology with RNA as a simple carrier molecule between DNA and proteins was overturned. Today RNA is recognized as an active player in biology, both as naturally occurring RNA enzymes or ribozyme, including the ribosome and likely the spliceosome, These findings, and the fundamental ability of RNA to act both as an efficient information carrier and functional macromolecule have led to wide speculation of the existence of an RNA World early in evolution.
With the discovery nearly thirty years ago that RNA can catalyze reactions with proficiencies that approach those of protein enzymes, the central dogma of biology with RNA as a simple carrier molecule between DNA and proteins was overturned. Today RNA is recognized as an active player in biology, both as naturally occurring RNA enzymes or ribozyme, including the ribosome and likely the spliceosome, These findings, and the fundamental ability of RNA to act both as an efficient information carrier and functional macromolecule have led to wide speculation of the existence of an RNA World early in evolution.
Thus, we explore RNA catalysis for several reasons: to learn more about the catalytic potential of RNA, how it evolved, and how it may someday be co-opted for medical, technological and industrial processes; to learn about the RNA molecules that function in modern-day biology and define its capabilities and limitations; and to provide a counterpoint to the vast knowledge of protein enzymes in order to help decipher what is fundamental to biological catalysts.
We are currently focused on the group I ribozyme, the most well-studied catalytic RNA in both structure and function. We harness previous studies, including multiple crystal structures, a robust phylogeny model, and a defined kinetic and thermodynamic framework for the Tetrahymena group I ribozyme, to delve more deeply into questions about catalytic RNA and how an RNA scaffold can be used to sculpt an active site. Some questions of interest include: What are the structural and functional roles of metal ions within the ribozyme, both inside and outside of the active site? What are the ways in which long-range interactions, distant from the active site, establish the active site and affect structure and activity? In what manner do specific hydrogen bonds modulate ribozyme function throughout the course of the reaction? We are also very interested in RNA conformational changes, as these transitions are key elements in nearly all RNA-mediated events.
To answer the aforementioned and additional questions, we use techniques including site-directed mutagenesis and site-specific chemical modifications to alter both the ribozyme itself and its substrates. The replacement of single functional groups within a complex RNA structure with multiple related functionalities is straightforward, whereas the corresponding replacements in proteins remains highly challenging. Function is probed via pre-steady state kinetics, and structure is probed using a battery of chemical footprinting approaches. Recent advances allow us to incorporate probes of local dynamics and single molecule fluorescence assays of functional conformational transitions.
Some leading papers from the lab in the area of RNA catalysis are:
- Herschlag, D. and Cech, T.R. (1990) Biochemistry 29, 10159-10171. “Catalysis of RNA Cleavage by the Tetrahymena thermophila Ribozyme. 1. Kinetic Description of the Reaction of an RNA Substrate Complementary to the Active Site.”(Medline) (PDF File)
- Narlikar, G.J., Gopalakrishnan, V., McConnell, T.S., Usman, N. and Herschlag, D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3668-3672. “Use of Binding Energy by an RNA Enzyme for Catalysis by Positioning and Substrate Destabilization.” (Medline) (PDF File)
- Peracchi, A., Beigelman, L., Usman, N. and Herschlag, D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11522-11527. “Rescue of Abasic Hammerhead Ribozymes by Exogenous Addition of Specific Bases.” (Medline) (PDF File)
- Hertel, K.J., Peracchi, A., Uhlenbeck, O.C. and Herschlag, D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8497-8502. “Use of Intrinsic Binding Energy for Catalysis by an RNA Enzyme.”(Medline) (PDF File)
- Narlikar, G.J. and Herschlag, D. (1998) Biochemistry 37, 9902-9911. “Direct Demonstration of the Catalytic Role of Binding Interactions in Enzymatic Reactions.” (Medline) (PDF File)
- Wang, S., Karbstein, K., Peracchi, A., Beigelman, L. and Herschlag, D. (1999) Biochemistry 43, 14363-14278. “Identification of the Hammerhead Ribozyme Metal Ion Binding Site Responsible for Rescue of the Deleterious Effect of a Cleavage Site Phosphorothioate.” (Medline) (PDF File)
- Shan, S., Kravchuk, A.V., Piccirilli, J.A. and Herschlag, D. (2001) Biochemistry 40, 5161-5171. “Defining the Catalytic Metal Ion Interactions in the Tetrahymena Ribozyme Reaction.” (Medline) (PDF File)
- Karbstein, K. and Herschlag, D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2300-2305. “Extraordinarily Slow Binding of Guanosine to the Tetrahymena Group 1 Ribozyme.” (Medline) (PDF File)
- Bartley, L.E., Zhuang, X., Das, R., Chu, S. and Herschlag, D. (2003) J. Mol. Biol. 328, 1011-1026. “Exploration of the Transition State for Tertiary Structure Formation between an RNA Helix and a Large Structured RNA.” (Medline) (PDF File)
- Forconi, M., Lee, J., Lee, J., Piccirilli, J., and Herschlag, D. (2008) Biochemistry 47, 6883-6894 “Functional Identification of Ligands for a Catalytic Metal Ion in Group I Introns.” (Medline) (PDF File
-
Grant, G.P.G., Boyd, N., Herschlag, D. and Qin, P.Z. (2009) J. Am. Chem. Soc. 131, 3136–3137. “Motions of the Substrate Recognition Duplex in a Group I Intron Assessed by Site-Directed Spin-Labeling.” (Medline) (PDF File)
See also: Faculty of 1000 Biology, Feb 2009 http://www.f1000biology.com/article/id/1156905/evaluation - Shi, X., Mollova, E., Pljevaljcic, G., Millar, D. and Herschlag, D. (2009) J. Am. Chem. Soc. 131, 9571-9578. “Probing the Dynamics of the P1 Helix within the Tetrahymena Group I Intron.” (Medline) (PDF File)
Useful reviews in the area of RNA catalysis are:
- Narlikar, G.J. and Herschlag, D. (1997) Annu. Rev. Biochem. 66,19-59. “Mechanistic Aspects of Enzymatic Catalysis: Lessons from Comparison of RNA and Protein Enzymes.” (Medline) (PDF File)
- Hougland, J., Piccirilli, J., Forconi, M., Lee, J. and Herschlag, D. (2005) RNA World 3rd Edition “How the Group I Intron Works: A Case Study of RNA Structure and Function.” Gesteland, R.F., Cech, T.R. and Atkins, J.F., Editors. Cold Spring Harbor Laboratory Press, New York. pp. 133-205. (PDF File)
RNA Folding
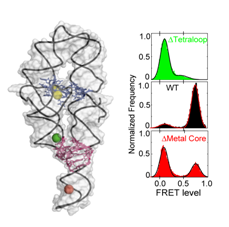 The repertoire of biological functions of RNA goes well beyond that of an information-carrying intermediary between DNA and proteins. RNA plays roles in a broad variety of cellular processes. In many cases, functional proficiency of RNA molecules is tightly linked to their ability to fold into specific, active conformations. Indeed, the molecular constituents of RNA render it highly prone to folding into highly stable structures. This tendency may have aided RNA as the central informational and functional molecule early in evolution in an 'RNA World'. In modern-day biology, this tendency of RNA to form stable structures likely led to the evolution and preponderance of RNA chaperones –i.e., proteins that help RNA unfold, explore its conformational space, and undergo functional conformational changes.
The repertoire of biological functions of RNA goes well beyond that of an information-carrying intermediary between DNA and proteins. RNA plays roles in a broad variety of cellular processes. In many cases, functional proficiency of RNA molecules is tightly linked to their ability to fold into specific, active conformations. Indeed, the molecular constituents of RNA render it highly prone to folding into highly stable structures. This tendency may have aided RNA as the central informational and functional molecule early in evolution in an 'RNA World'. In modern-day biology, this tendency of RNA to form stable structures likely led to the evolution and preponderance of RNA chaperones –i.e., proteins that help RNA unfold, explore its conformational space, and undergo functional conformational changes.
In the Herschlag lab, we study RNA folding at all level, from complex functional RNAs that reveal the behavior of molecules with functional biological roles, to simple model systems that allow us to interrogate the energetics that underlie the folding and behavior of RNA molecules. The former approach has required the use of cutting edge technologies, including single molecule fluorescence, small angle X-ray scattering, EPR, and the development of new dynamics and and other methods. The latter approach has provided some of the first critical tests of theory and provides guides for the develop of new computational approaches.
Most generally, we can view RNA folding as modular, and this modularity greatly simplifies the process, facilitates energetic and conceptual dissection, and allows us to break down complex behaviors into simple physical concepts. RNA secondary structure, double-stranded helices, are generally stable; the forces between them are largely electrostatic and greatly modulated by the 'ion atmosphere' that surrounds all polyelectrolytes. These helices are linked by junctions, and we are exploring the conformational behavior of these junctions and how they bias the conformational space of the attached helices and thus affect the folding stability and specificity; this work has analogy to the development of the Ramachandran plot for peptide conformational preferences. Finally, RNA is linked together in tertiary space by motifs, RNA structural units that can be used multiple times within a folded RNA and are used common to many different folded RNAs. Our goal is to reach a physics-based level of description of RNA folding landscapes. We believe that such level of understanding of macromolecular behavior is a necessary step towards understanding the workings of complex biological machines, and, eventually, the process of life.
Like other projects, our work in RNA folding is highly interdisciplinary, involving biochemistry, molecular biology, chemistry, and physics. Some leading papers from the lab in the area of RNA folding are:
- Zhuang, X., Bartley, L.E., Babcock, H.P., Russell, R., Ha, T., Herschlag, D. and Chu, S. (2000) Science 288, 2048-2051. "A Single Molecule Study of RNA Catalysis and Folding." (Medline) (PDF File)
- Russell, R., Zhaung, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S. and Herschlag, D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 155-160. "Exploring the Folding Landscape of a Structured RNA." (Medline) (PDF File)
- Russell, R., Millett, I.S., Tate, M.W., Kwok, L.W., Nakatani, B., Gruner, S.M., Mochrie, S.G.J., Pande, V., Doniach, S., Herschlag, D. and Pollack, L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4266-4271. "Rapid Compaction During RNA Folding." (Medline) (PDF File)
- Das, R., Mills, T.T., Kwok, L.W., Maskel, G.S., Millett, I.S., Doniach, S., Finkelstein, K.D., Herschlag, D. and Pollack, L. (2003) Physical Review Letters 90, 188103-1 - 188103-4. "The Counterion Distribution Around DNA Probed by Solution X-Ray Scattering." (Medline) (PDF File)
- Takamoto, K., Das, R., He, Q., Doniach, S., Brenowitz, M., Herschlag, D. and Chance, M. (2004) J. Mol. Biol. 343, 1195-1206. "Principles of RNA Compaction: Insights From the Equilibrium Folding Pathway of the P4-P6 RNA Domain in Monovalent Cations." (Medline) (PDF File)
- Bai, Y., Das, R., Millet, I.S., Herschlag, D. and Doniach, S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1035-1040. "Probing Counterion Modulated Repulsion and Attraction Between Nucleic Acid Duplexes in Solution." (Medline) (PDF File)
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 6190-6195 "Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins." (Medline) (PDF File)
- Bai, Y., Travers, K., Chu, V.B., Lipfert, J., Doniach, S. and Herschlag, D. (2007) J. Am. Chem. Soc. 29, 14981-14988. " Quantitative and Comprehensive Decomposition of the Ion Atmosphere around Nucleic Acids." (Medline) (PDF File)
- Shi, X., Mollova, E., Pljevaljcic, G., Millar, D. and Herschlag, D. (2009) J. Am. Chem. Soc. 131, 9571-9578. "Probing the Dynamics of the P1 Helix within the Tetrahymena Group I Intron." (Medline) (PDF File)
- Zalatan, J.G. and Herschlag, D. (2009) Nature Chem. Biol. 5, 516-520. "The Far Reaches of Enzymology." PMCID in process (Medline) (PDF File)
Useful reviews in the area of RNA folding are:
- Herschlag, D. (1995) J. Biol. Chem. 270, 20871-20874. "RNA Chaperones and the RNA Folding Problem."(Medline) (PDF File)
- Chu, V.B. and Herschlag, D. (2008) Curr. Opin. Struc. Biol. 18, 305-314. "Unwinding RNA's Secrets: Advances in the Biology, Physics, and Modeling of Complex RNAs." (Medline) (PDF File)
- Chu, V.B., Bai, Y., Lipfert, Y., Doniach, S. and Herschlag, D. (2008) Curr. Opin. Chem. Biol. 12, 619-625. "A Repulsive Field: Advances in the Electrostatics of the Ion Atmosphere." (Medline) (PDF File)