Chaos
CHAOS
FUNCTION
Chaos makes a CHAOS game representation of a nucleic acid sequence using the method of Jeffrey (1990) Nucleic Acids Research 18: 2163-2170.
DESCRIPTION
Chaos plots a GCG nucleotide sequence in CHAOS game representation format, using the method of Jeffrey (1990). This method gives a display of a gene sequence which displays both local and global patterns.
The plot can show under represented sequence motifs, such as CG in human sequences or CATG in E. coli. It can also show overrepresented sequences in many cases.
AUTHOR
This program was written by Rodrigo Lopez S. (E-mail: rodrigol@biotek.uio.no; Post: Biotechnology Centre of Oslo, PO Box 1125 Blindern, N-0317 Oslo 3, Norway).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a sample session of Chaos
% chaos
CHAOS uses DNA sequences
CHAOS of what sequence ? GenEMBL:Hsambptr
. . EMPR:HSAMBPTR
Start (* 1 *) ?
End (* 5022*) ?
%
INPUT FILE
The input file for Chaos is a GCG nucleotide sequence file.
OUTPUT FILE
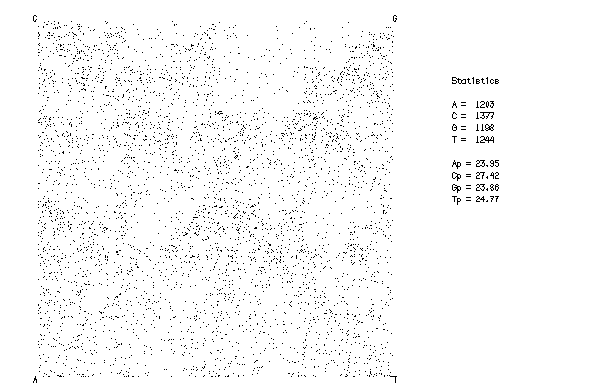
This is the plot from the example session
ALGORITHM
Chaos plots one point for each base in the sequence, according to a very simple rule.
The plot region is a square, with one base marked in each corner. Imagine that the pen starts in the centre of the square. For each base, starting at the beginning of the sequence, the pen moves half way to that base's corner of the plot and draws a dot. This continues until the end of the last sequence is reached.
The method is very simple, but the results can show great detail about the sequence. Because of the way the plot is drawn, even though the points appear to be scattered, the point for a G is always drawn in the top right quarter of the plot. Exactly where the point is will depend on where the pen was for the previous base. For example, if it was a C the pen would have been in the top left quarter of the plot.
Combining these two, any G which comes after a C will be plotted in the same 1/sixteenth square, just to the right of centre at the top of the plot.
The example plot shows a human sequence of about 5kb, just long enough to show features. For most human sequences, the sequence CG is rare (see also the CpGPlot program). This is shown by the reduction in the number of dots in this region.
Other, smaller, square regions with fewer dots can also be seen. These correspond to longer sequences which include CG (ACG, CCG, GCG, TCG, AACG and so on). Given enough sequence data, detail can be seen down to the resolution of the plot. For example, a plot of one of the long E.coli genomic sequences (for example ECUW87, accession number L19201) will clearly show details of pentanucleotide sequence variation (Merkl et al, 1992).
COMMAND-LINE SUMMARY
All parameters for this program may be put on the command line. Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes. In the summary below, the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter. Square brackets ([ and ]) enclose qualifiers or parameter values that are optional. For more information, see "Using Program Parameters" in Chapter 3, Basic Concepts: Using Programs in the GCG User's Guide.
Minimum syntax: % chaos [-INfile=]empri:hsldlr -Default Prompted Parameters: -BEGin=1 -END=1000 the range of interest -RNA the sequence is RNA Local Data Files: None Optional Parameters: None
OPTIONAL PARAMETERS
None.
REFERENCES
Jeffrey, H.J. (1990). Chaos game representation of gene structure. Nucleic Acids Research 18, 2163-2170.
Merkl, R., Kroeger, M., Rice, P., Fritz, H-J. (1992). Statistical evaluation and biological interpretation of non-random abundance in the E.coli K-12 genome of tetra- and pentanucleotide sequences related to VSP DNA mismatch repair. Nucleic Acids Research 20, 1657-1662.
Printed: April 22, 1996 15:52 (1162)