Clustree
CLUSTREE(+)
FUNCTION
ClusTree computes a phylogenetic tree according to the Neighbor-Joining Method of Saitou and Nei (1987). This is part of the original ClustalW distribution, modified for inclusion in EGCG. The tree will be displayed graphically.
DESCRIPTION
ClusTree allows you to input an alignment and calculate a phylogenetic tree. The sequences must be aligned already! The tree will look strange if the sequences are not already aligned. You can also "BOOTSTRAP" the tree to show confidence levels for groupings. The method used is the Neighbor-Joining method of Saitou and Nei (Mol. Biol. Evol. 4: 406-425 (1987)). This is a "distance method". First, percent divergence figures are calculated between all pairs of sequences. These divergence figures are then used by the NJ method to give the tree.
AUTHOR
ClustalW was written by Des Higgins (E-mail:Des.Higgins@ebi.ac.uk)
The EGCG version of the program was modified by Weiyun Chen and Karl-Heinz Glatting at the German Cancer Research Centre (DKFZ), Heidelberg, Germany.
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
% clustree
clustree of what profile ? globin.msf{}
What should I call the tree output file (* globin.nj *) ?
%
OUTPUT
Here is the output file:
clustree August 16, 1995 16:58
of: globin.msf{}
Phylogenetic tree parameters:
Exclude positions with gaps: No
Correct for multiple substitutions: No
DIST = percentage divergence (/100)
Length = number of sites used in comparison
1 vs. 2 DIST = 0.5612; length = 139
1 vs. 3 DIST = 0.5899; length = 139
1 vs. 4 DIST = 0.7557; length = 131
1 vs. 5 DIST = 0.6489; length = 131
1 vs. 6 DIST = 0.7857; length = 140
1 vs. 7 DIST = 0.7305; length = 141
2 vs. 3 DIST = 0.2671; length = 146
2 vs. 4 DIST = 0.7612; length = 134
2 vs. 5 DIST = 0.7500; length = 136
2 vs. 6 DIST = 0.8112; length = 143
2 vs. 7 DIST = 0.7517; length = 145
3 vs. 4 DIST = 0.7388; length = 134
3 vs. 5 DIST = 0.7206; length = 136
3 vs. 6 DIST = 0.8042; length = 143
3 vs. 7 DIST = 0.7586; length = 145
4 vs. 5 DIST = 0.5753; length = 146
4 vs. 6 DIST = 0.8175; length = 137
4 vs. 7 DIST = 0.8235; length = 136
5 vs. 6 DIST = 0.8261; length = 138
5 vs. 7 DIST = 0.7810; length = 137
6 vs. 7 DIST = 0.8138; length = 145
Neighbor-Joining Method
Saitou, N. and Nei, M. (1987) The Neighbor-Joining Method:
A New Method for Reconstructing Phylogenetic Trees.
Mol. Biol. Evol., 4(4), 406-425
This is an UNROOTED tree
Numbers in parentheses are branch lengths
Cycle 1 = SEQ: 2 ( 0.13587) joins SEQ: 3 ( 0.13125)
Cycle 2 = SEQ: 4 ( 0.30711) joins SEQ: 5 ( 0.26823)
Cycle 3 = SEQ: 1 ( 0.27327) joins Node: 2 ( 0.16871)
Cycle 4 = Node: 1 ( 0.05233) joins Node: 4 ( 0.09470)
Cycle 5 (Last cycle, trichotomy):
Node: 1 ( 0.02967) joins
SEQ: 6 ( 0.41835) joins
SEQ: 7 ( 0.38165)
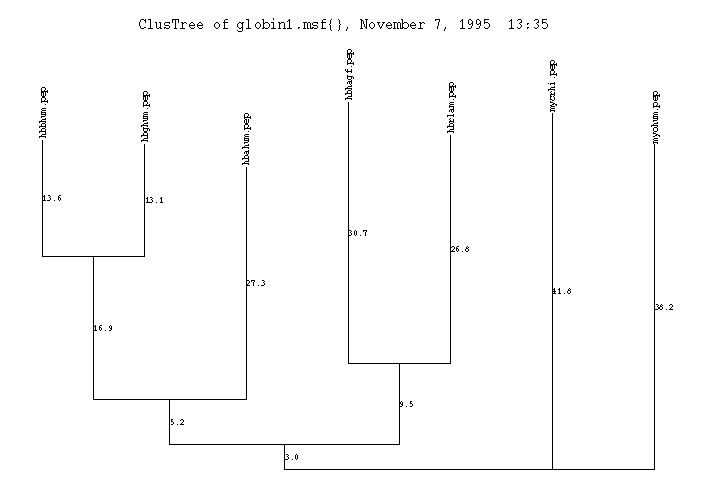
This is the plot from the example session.
RELATED PROGRAMS
BoxAlign displays a sequence alignment graphically marking columns with conserved amino-acids or nucleotides with boxes. BoxAlign does not compute an alignment, it simply displays it.
EClustAlW calculates a multiple alignment of nucleic acid or protein sequences according to the method of Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994). This is part of the original ClustalW distribution, modified for inclusion in EGCG.
LineUp is a screen editor for editing multiple sequence alignments. You can edit up to 30 sequences simultaneously. New sequences can be typed in by hand or added from existing sequence files. A consensus sequence identifies places where the sequences are in conflict.
MultAlign does a simultaneous alignment for two or more DNA or protein sequences. It introduces a certain number of gaps into either pairwise aligned sequences or groups of sequences to find a minimal global distance. The user can influence the result by defining the order in which the sequences will be aligned. The program is based on a generalization of the algorithm of Waterman, Smith and Beyer by Krueger and Osterburg.
Motifs looks for sequence motifs by searching through proteins for the patterns defined in the PROSITE Dictionary of Protein Sites and Patterns. Motifs can display an abstract of the current literature on each of the motifs it finds.
MultAlign does a simultaneous alignment for two or more DNA or protein sequences. It introduces a certain number of gaps into either pairwise aligned sequences or groups of sequences to find a minimal global distance. The user can influence the result by defining the order in which the sequences will be aligned. The program is based on a generalization of the algorithm of Waterman, Smith and Beyer by Krueger and Osterburg.
PileUp creates a multiple sequence alignment from a group of related sequences using progressive, pairwise alignments. It can also plot a tree showing the clustering relationships used to create the alignment.
PlotAlign takes a GCG format sequence alignment, and plots the mean and range of values for any amino acid parameter you supply. The "panel file" contains a list of parameters to be plotted. The main database of parameters is taken from Nakai et al. (1988), and the default panel file uses selected parameters from the 13 discrete clusters in that paper. This program is experimental. Any suggestions would be most welcome.
Pretty displays multiple sequence alignments and calculates a consensus sequence. It does not create the alignment; it simply displays it.
ProfAlign is for taking two old aligments (or single sequences) and aligning them with each other. The result is one bigger aligment. This is part of the original ClustalW distribution, modified for inclusion in EGCG.
ProfileGap makes an optimal alignment between a profile and a sequence.
TProfileGap makes an optimal alignment between a profile and a sequence.
Tree produces a multiple alignment for a set of protein sequences by iteratively acting on the sequences. An approximate phylogenetic order of the sequences is first determinded by a series of pairwise alignments using the Needleman and Wunsch method. Any subclusters that may exist in the set are prealigned before the final alignment is undertaken. Finally, the phylogenetic tree of the sequences is plotted in the form of a dendrogram.
GRAPHICS
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
CTRL-C
If you need to stop this program,
use By default,
ClusTree writes instruction for plotting the tree into a figure file named clustree.figure.
Such file can be plotted on any supported graphics device using the Figure
program.
The sequences included in the alignment you use for running ClusTree must not exceed the total number of 500 while each sequence used can consist of up to 10,000 symbols.
It has to be considered that as gaps are inserted in the alignment,
the length in the final alignment cannot exceed 10,000 characters for any sequence.
This means,
the maximum sequence length is 10,000-X,
where X is the number of gaps introduced by the program that creates the alignment.
The phylogenetic trees (the real trees that you calculate AFTER alignment;
not the guide trees used to decide the branching order for multiple alignment)
use the Neighbor-Joining method of Saitou and Nei (1987)
based on a matrix of "distances" between all sequences.
These distances can be corrected for "multiple hits".
This is normal practice when accurate trees are needed.
This correction stretches distances (especially large ones)
to try to correct for the fact that OBSERVED distances (mean number of differences per site)
greatly underestimate the actual number that happened during evolution.
The formula used to correct for multiple hits is from Motoo Kimura (In The Neutral Theory of Molecular Evolution,
p.
75,
Cambridge University Press,
Cambridge,
England,
1983)
and is: K = -Ln(1 - D - (D.D)/5)
where D is the observed distance and K is corrected distance.
This formula gives mean number of estimated substitutions per site and,
in contrast to D (the observed number),
can be greater than 1 i.e.
more than one substitution per site,
on average.
For example,
if you observe 0.8 differences per site (80% difference;
20% identity),
then the above formula predicts that there have been 2.5 substitutions per site over the course of evolution since the 2 sequences diverged.
This can also be expressed in PAM units by multiplying by 100 (mean number of substitutions per 100 residues).
The PAM scale of evolution and its derivation/calculation comes from the work of Margaret Dayhoff and co workers (the famous Dayhoff PAM series of weight matrices also came from this work).
Dayhoff et al constructed an elaborate model of protein evolution based on observed frequencies of substitution between very closely related proteins.
Using this model,
they derived a table relating observed distances to predicted PAM distances.
Kimura's formula,
above,
is just a "curve fitting" approximation to this table.
It is very accurate in the range 0.75 > D > 0.0 but becomes increasingly inaccurate at high D (>0.75)
and fails completely at around D = 0.85.
To circumvent this problem,
all the values for K corresponding to D above 0.75 are calculated directly using the Dayhoff model and these are stored in an internal table,
used by EClustalW.
This table gives values of K for all D between 0.75 and 0.93 in intervals of 0.001 i.e.
for D = 0.750,
0.751,
0.752 ......
0.929,
0.930.
For any observed D higher than 0.930,
we arbitrarily set K to 10.0.
This sounds drastic but with real sequences,
distances of 0.93 (less than 7% identity)
are rare.
If your data set includes sequences with this degree of divergence,
you will have difficulty getting accurate trees by ANY method;
the alignment itself will be very difficult (to construct and to evaluate).
There are some important things to note.
Firstly,
this formula works well if your sequences are of average amino acid composition and if the amino acids substitute according to the original Dayhoff model.
In other cases,
it may be misleading.
Secondly,
it is based only on observed percent distance i.e.
it does not DIRECTLY take conservative substitutions into account.
Thirdly,
the error on the estimated PAM distances may be VERY great for high distances;
at very high distance (e.g.
over 85%)
it may give largely arbitrary corrected distances.
In most cases,
however,
the correction is still worth using;
the trees will be more accurate and the branch lengths will be more realistic.
A far more sophisticated distance correction based on a full Dayhoff model which DOES take conservative substitutions and actual amino acid composition into account,
may be found in the PROTDIST program of the PHYLIP package.
For serious tree makers,
this program is highly recommended.
When you use the BOOTSTRAP in EClustalW
to estimate the reliability of parts of a tree,
many of the uncorrected distances may randomly exceed the arbitrary cut off of 0.93 (sequences only 7% identical)
if the sequences are distantly related.
This will happen randomly i.e.
even if none of the pairs of sequences are less than 7% identical,
the bootstrap samples may contain pairs of sequences that do exceed this cut off.
If this happens,
you will be warned.
In practice,
this can happen with many data sets.
It is not a serious problem if it happens rarely.
If it does happen (you are warned when it happens and told how often the problem occurs),
you should consider removing the most distantly related sequences and/or using the PHYLIP package instead.
A further problem arises in almost exactly the opposite situation: when you bootstrap a data set which contains 3 or more sequences that are identical or almost identical.
Here,
the sets of identical sequences should be shown as a multifurcation (several sequences joing at the same part of the tree).
Because the Neighbor-Joining method only gives strictly dichotomous trees (never more than 2 sequences join at one time),
this cannot be exactly represented.
In practice,
this is NOT a problem as there will be some internal branches of zero length seperating the sequences.
If you display the tree with all branch lengths,
you will still see a multifurcation.
However,
when you bootstrap the tree,
only the branching orders are stored and counted.
In the case of multifurcations,
the exact branching order is arbitrary but the program will always get the same branching order,
depending only on the input order of the sequences.
In practice,
this is only a problem in situations where you have a set of sequences where all of them are VERY similar.
In this case,
you can find very high support for some groupings which will disappear if you run the analysis with a different input order.
Again,
the PHYLIP package deals with this by offering a JUMBLE option to shuffle the input order of your sequences between each bootstrap sample.
All parameters for this program may be put on the command line.
Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes.
In the summary below,
the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter.
Square brackets ([ and ])
enclose qualifiers or parameter values that are optional.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
For details about the ClustAlW program package,
including ClustAl,
ProfAlign
and Clustree,
see J.
D.
Thompson et al.
(Nucleic Acids Research,
22 (22): 4673-4680 (1994))
and D.
G.
Higgins et al.
(CABIOS 8 (2):189-191 (1992)).
For details about the overall multiple alignment algorithm see D.
G.
Higgins and P.
M.
Sharp (CABIOS 5: 151-153 (1989)).
ClusTree is part of ClustalW which was developped and written by Des Higgins,
European BioInformatics Institute,
EMBL Outstation,
Hinxton,
UK.
The program was added to the Package for HUSAR version 3.0 by Weiyun Chen and Karl-Heinz Glatting,
DKFZ Heidelberg,
Germany,
and converted to EGCG by Peter Rice,
Sanger Centre,
Hunxton,
UK.
None
The parameters and switches listed below can be set from the command line.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
Supresses the plot of the tree.
As sequences diverge,
substitutions accumulate.
The parameter -KIMURA
corrects the calculation of the phylogenetic tree in case there were several substitutions at the one site,
but only one of them observe.
It stretches the long branches of the tree while leaving the short ones relatively untouched.
The desired effect is to try to make distances proportional to time since divergence.
The optional parameter -TOSSGAPS
allows you to ignore all alignment positions (columns)
where there is a gap in any sequence.
This guarantees that "like" is compared with "like" in all distances i.e.
the same positions are used to calculate all distances.
It also means that the distances will be "metric".
The disadvantage of using this option is that you throw away much of the data if there are many gaps.
If the total number of gaps is small,
it has little effect.
The parameter -BOOTstrap provides a method called bootstrapping to give you an indication of the degree of error in the phylogenetic tree.
The seed number for the random number generator.
Different bootstrapping runs with the same seed number will give the same answer.
If you wish to carry out genuinely different bootstrap sampling experiments,
give different seed numbers.
These options apply to all GCG graphics programs.
These and many others are described in detail in Chapter 5,
Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure
program instead of drawing the plot on your plotter.
draws all text characters on the plot using Font 3 (see Appendix I)
.
draws the entire plot with the pen in stall 1.
These options let you expand or reduce the plot (zoom),
move it in either direction (pan),
or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0)
to 1.2 (zoom in).
You can expand the axes independently with -XSCAle and -YSCAle.
Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees.
Usually,
plots are displayed with the horizontal axis longer than the vertical (landscape).
Note that plots are reduced or enlarged,
depending on the platen size,
to fill the page.
Saitou N.
and Nei M.
(1987).
"The neighbor-joining method: a new method for reconstructing phylogenetic trees." Mol.
Biol.
Evol.
4,
406-425.
Thompson J.D.,
Higgins D.G.
and Gibson T.J.
(1994)
"CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting,
position-specific gap penalties and weight matrix choice." Nucleic Acids Research 22,
4673-4680.
Higgins D.G.,
Bleasby A.J.
and Fuchs R.
(1992).
"CLUSTAL V: improved software for multiple sequence alignment." Comput.
Appl.
Biosci.
8,
189-191.
Higgins D.G.
and Sharp P.M.
(1989).
"Fast and sensitive multiple sequence alignments on a microcomputer." Comput.
Appl.
Biosci.
5,
151-153.
Printed: April 22,
1996 15:52 (1162)
Here is the plot of the example session:
RESTRICTIONS
ALGORITHM
Bootstrap
COMMAND-LINE SUMMARY
Minimal Syntax: % clustree [-PROFile=]globin.msf{*} -Default
Prompted Parameters:
[-OUTfile=]globin.nj output file name
Optional Parameters:
-NOPLOT do not plot the tree
-KIMURA correct for multiple substitutions
-TOSSGAPS exclude positions with gaps
-BOOTstrap[=1000] number of bootstrapping samples
-SEED=111 seed number for random number generator
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
ACKNOWLEDGEMENT
LOCAL DATA FILES
OPTIONAL PARAMETERS
-NOPLOT
-KIMURA
-TOSSGAPS
-BOOTstrap[=1000]
-SEED=111
-FIGure=programname.figure
-FONT=3
-COLor=1
-SCAle=1.2
-XPAN=30.0
-YPAN=30.0
-PORtrait
REFERENCES