Quickmatch
- QUICKMATCH(+)
- FUNCTION
- DESCRIPTION
- AUTHOR
- EXAMPLE
- OUTPUT
- RELATED PROGRAMS
- RESTRICTIONS
- ALGORITHM
- CONSIDERATIONS
- INPUT FILES
- GRAPHICS
- CTRL-C
- COMMAND-LINE SUMMARY
- FUTURE DEVELOPMENT
- ACKNOWLEDGEMENTS
- LOCAL DATA FILES
- OPTIONAL PARAMETERS
- -PLOt
- -RANdom
- -WORd
- -CAPtion
- -ALL
- -TICKAXes
- -DOTSonly
- -NOSHOWalign
- -NOSELF
- -PERFect
- -STRingency
- -WHOle Performs a full Needleman and Wunsch alignment (like Gap) rather than
- -LIMit
- -GAPweight
- -LENgthweight
- -PAIr
- -PAGe
- -WIDth
- -NOBIGGaps
- -MONitor
- -SUMmary
- -DENsity
- -FIGure
- -FONT
- -COLor
- -SCAle
- -XPAN
- -YPAN
- -PORtrait
QUICKMATCH(+)
FUNCTION
QuickMatch displays the overlaps found by EQuickSearch with either optimal alignments or dot-plots. The alignments can be selected by quality to discard poor matches. The dot-plots can be reviewed rapidly with a graphic screen.
NOTE: The EGCG Quick Searching System programs are now fully supported by the EGCG team. GCG distributed the original programs in the hope that users would make suggestions about their future development. This program is one such suggestion.
DESCRIPTION
EQuickSearch identifies overlaps between a query sequence and a sequence database. QuickMatch displays those overlaps as either dot-plots or optimal alignments.
If you have a fast graphics device (such as a Postscript printer), the dot-plots are a powerful way to see the nature of the overlap. If you don't, optimal alignments will have to do.
QuickMatch is a modified version of the original GCG program QuickShow. Both programs read the same input file, and produce the same graphics output. The changes are in various command line options to allow selection of only those alignments that are perfect, or that are above a specified quality.
QuickMatch by default behaves in the same way as QuickShow, except that the menu defaults to alignments rather than dot plots, and the output file name defaults to ".match" rather than ".quickshow".
AUTHOR
This program was written by Peter Rice (E-mail: pmr@sanger.ac.uk Post: Informatics Division, The Sanger Centre, Hinxton Hall, Cambridge, CB10 1RQ, UK).
All EGCG programs are supported by the EGCG Support Team, who can be contacted by E-mail (egcg@embnet.org).
EXAMPLE
Here is a session with QuickMatch that was used to display the overlaps between ggammacod.seq (a human fetal beta globin G-gamma coding sequence) and the GenEMBL database. The input file is the output from the example session for EQuickSearch.
% quickmatch -STRingency=0.95 -WHOle
QUICKMATCH of what file ? ggammacod.quick
Display the overlaps with:
1) dot plots or
2) optimal alignments?
3) list of accepted hits
Please choose one (* 2 *) :
What should I call the output file (* ggammacod.match *) ?
Query "+": ggammacod.seq Len: 444 from: 1 to: 444
Overlap: Ggagglog Len: 1,797 from: 1 to: 1,797
///////////////////
There were 19 comparisons.
Stringency: 0.95
Accepted: 16
Rejected: 3
%
OUTPUT

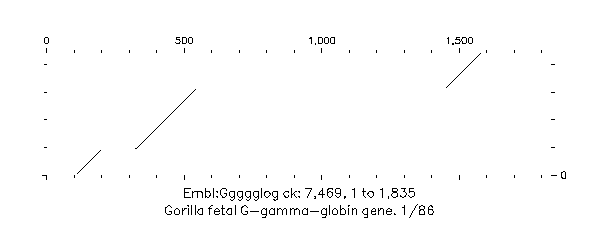
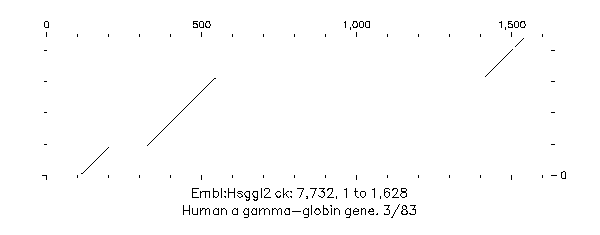
The first three dot-plots from this input file are shown in Figures above. In each dot-plot the query sequence ggammacod.seq is on the vertical axis, while the various overlapping sequences found by EQuickSearch are on the horizontal.
If you had chosen to display the overlaps with optimal alignments, QuickMatch would have written a file like this one:
(BestFit) QUICKMATCH of: Ggammacod.Quick March 31, 1990 15:22
** Stringency: 0.95 **
! QUICKSEARCH of: Gendocdata:Ggammacod.Seq March 13, 1990 15:17
Comparison Table: Gencoredisk:[Gcgcore.Rundata]Quickdna.Cmp
Gap Weight: 5.00 Gap Length Weight: 0.10 ..
Ggammacod.Seq Check: 2,906 length: 444 from: 1 to: 444
Coding sequence for Human fetal beta globin G-gamma.
Ggagglog Check: 7,760 length: 1,797 from: 1 to: 1,797
Gorilla fetal A-gamma-globin gene. 1/86
Diagonal: 229 Range: -443/+444
Gaps: 0 Quality: 221.3 Ratio: 0.975
. . . . .
91 AGGCTCCTGGTTGTCTACCCATGGACCCAGAGGTTCTTTGACAGCTTTGG 140
||||||||||||||||||||||||||||||||||||||||||||||||||
320 AGGCTCCTGGTTGTCTACCCATGGACCCAGAGGTTCTTTGACAGCTTTGG 369
. . . . .
141 CAACCTGTCCTCTGCCTCTGCCATCATGGGCAACCCCAAAGTCAAGGCAC 190
||||||||||||||||||||||||||||||||||||||||||||||||||
370 CAACCTGTCCTCTGCCTCTGCCATCATGGGCAACCCCAAAGTCAAGGCAC 419
. . . . .
191 ATGGCAAGAAGGTGCTGACTTCCTTGGGAGATGCCATAAAGCACCTGGAT 240
|||||||||||||||||||||||||||||| |||||||||||||||||||
420 ATGGCAAGAAGGTGCTGACTTCCTTGGGAGGTGCCATAAAGCACCTGGAT 469
. . . . .
241 GATCTCAAGGGCACCTTTGCCCAGCTGAGTGAACTGCACTGTGACAAGCT 290
||||||||||||||||||||||||||||||||||||||||||||||||||
470 GATCTCAAGGGCACCTTTGCCCAGCTGAGTGAACTGCACTGTGACAAGCT 519
. .
291 GCATGTGGATCCTGAGAACTTCAAGCT 317
||||||||||||||||||||||| | |
520 GCATGTGGATCCTGAGAACTTCAGGGT 546
////////////////////////////////////////////
RELATED PROGRAMS
EQuickSearch rapidly identifies places where query sequence(s) occur in a nucleotide sequence database. The output is a file of overlaps that can be displayed with QuickMatch or EQuickShow. You can make up your own sequence database or use GenEMBL, which consists of GenBank and those sequences in EMBL that are not represented in GenBank (or the other way around at some sites).
RESTRICTIONS
There is no sequence length restriction when the -PERFect command line option is used. For other alignments, only the first 32,000 bases of the best diagonal can be aligned.
ALGORITHM
Dot Plots
The DotPlot option of QuickMatch is identical to the behaviour of the original QuickShow program.
QuickMatch reads the query sequence and the set of sequences to which it overlaps from the EQuickSearch overlap file. All the matching words of length eight generate a dot on the plot. To increase the speed of plotting, all the points that lie on diagonals with less than four times the expected number of random points are NOT displayed. Use the command line switch -RANdom if you want to see these points.
Alignments
First of all, just as with dot-plots, all the points between the overlapping sequences where there is a perfect match of eight base pairs are calculated. The diagonal with the most points is then identified.
For perfect alignments (with -PERFect on the command line), QuickMatch now simply checks the complete best diagonal, and displays the alignment if an exact match is found.
For all other alignments QuickMatch does a limited BestFit (or a Gap if the command line option -WHOle was used) on each pair of sequences to make an alignment near that diagonal. For a detailed description, see the descriptions of programs BestFit and Gap run with -LIMit on the command line. The gap limits are set to the maximum allowed by the alignment routines in the GCG Procedure Library, but you can set a smaller limit with the -LIMit command line option.
CONSIDERATIONS
QuickMatch was designed for a fast graphics device like a workstation, a Tektronix 4107, or a Graphon GO-250. QuickMatch takes about two seconds to calculate and display each plot on our Tektronix 4207 terminal. In the absence of such a terminal, a plotter with automatic page feeding like the Apple Laserwriter is a reasonable way to see the plots. Otherwise you are left with optimal alignments.
QuickMatch is supposed to help you recognize false positives quickly. Most of them arise when a word in the query hashes to a polymeric (simple repeat) sequence in the database.
In the alignments, the -PERFect and -STRingency command line options allow you to eliminate the false positives.
In dot-plots these polymeric sequences sometimes appear as a horizontal line of points instead of a diagonal. You can see these polymers more clearly if you put -RANdom on the command line.
GCG alignments display the back strand with descending top strand coordinates. GCG dot-plots, on the other hand, display the back strand with ascending coordinates that begin at the original strand's 3' end.
INPUT FILES
QuickMatch will read the file names in the output file from EQuickSearch. If any of the horizontal search set files have been changed or deleted, QuickMatch will act as if they do not exist. If the vertical query sequence cannot be read correctly, QuickMatch will complain. There are two back slashes '\\' in front of the query sequence and two forward slashes '/ -' at the end of the list of overlaps to that query. Here is the input file for the example session:
! EQUICKSEARCH of: Ggammacod.Seq February 1, 1989 18:41 ! WordSize: 20 Window: 15 Stringency: 7 .. \\ D22:[Burgess.Work]Ggammacod.Seq;1 Sgmt: 1 Strd: + ! Coding sequence for Human fetal beta globin G-gamma. !Sequence Sgmnt Words Documentation Primate:Chpagglog 1 18 !Chimpanzee fetal A-gamma-globin gene. Primate:Chpggglog 1 17 !Chimpanzee fetal G-gamma-globin gene. Primate:Humhbb 9 21 !Human beta globin region on chromosom Primate:Humhbb 10 14 !Human beta globin region on chromosome Primate:Humhbgab 1 16 !Human A-gamma-globin gene on chromosome Primate:Humhbgg 1 21 !Human glycine-gamma-globin, 3' end. Primate:Machbca2 1 17 !Rhesus monkey beta-cluster 5' fetal Primate:Machbga1 1 15 !Rhesus monkey beta-cluster 3' fetal Primate:Orahbg1f 1 19 !Orangutan gamma-1-fetal globin gene Primate:Orahbg2f 1 17 !Orangutan gamma-2-fetal globin gene Embl:Ggagglog 1 18 !Gorilla fetal A-gamma-globin gene. Embl:Ggggglog 1 20 !Gorilla fetal G-gamma-globin gene. Embl:Hsggl2 1 19 !Human a gamma-globin gene. 3/83 Embl:Hsggl3 1 16 !Human A-gamma-globin gene. 8/83 Embl:Hsggl4 1 21 !Human G-gamma-globin gene. 12/83 Embl:Hsglbn 1 20 !Human gene for fetal A-gamma and Embl:Hsglbn 2 14 !Human gene for fetal A-gamma and //
GRAPHICS
The Wisconsin Package must be configured for graphics before you run any program with graphics output! If the % setplot command is available in your installation, this is the easiest way to establish your graphics configuration, but you can also use commands like % postscript that correspond to the graphics languages the Wisconsin Package supports. See Chapter 5, Using Graphics in the User's Guide for more information about configuring your process for graphics.
CTRL-C
If you need to stop this program,
use
All parameters for this program may be put on the command line.
Use the option -CHEck to see the summary below and to have a chance to add things to the command line before the program executes.
In the summary below,
the capitalized letters in the qualifier names are the letters that you must type in order to use the parameter.
Square brackets ([ and ])
enclose qualifiers or parameter values that are optional.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
The usual GCG alignment options are available in QuickMatch The graphics output of QuickShow has not so far been changed.
Any suggestions for improvement would be welcome.
QuickMatch was written by Peter Rice at EMBL,
Heidelberg,
Germany.
It is a modification of the original program QuickShow which was designed by John Devereux and implemented by John Devereux and Christopher Dow of GCG,
and also includes alignment code from the GCG Segments
program.
The files described below supply auxiliary data to this program.
The program automatically reads them from a public data directory unless you either 1)
have a data file with exactly the same name in your current working directory;
or 2)
name a file on the command line with an expression like -DATa1=myfile.dat.
For more information see Chapter 4,
Using Data Files in the User's Guide.
For optimal alignments,
QuickMatch reads symbol comparison values from the file quickdna.cmp.
The parameters and switches listed below can be set from the command line.
For more information,
see "Using Program Parameters" in Chapter 3,
Basic Concepts: Using Programs in the GCG User's Guide.
suppresses the menu that presents the alternatives of either dot-plots or alignments and makes dot-plots.
Normally QuickMatch only plots points that lie on diagonals having eight times the number of points expected at random.
This option shows every point where there is a matching word regardless of the number of other points that occur on that diagonal.
The criterion for a point on the dot-plot is that there is a perfect match of 8 base pairs.
This option lets you select a word size between 1 and 20 if you don't like the default value of 8.
Smaller word comparisons are a little more sensitive,
but there are many more points in the background.
Adds a blue dividing box to the plot,
and some annotating text to its left.
When the sequences compared are identical in both range and checksum,
only the points above the diagonal are plotted.
The diagonal is represented with a line.
You can override this feature with the command line switch -ALL.
GCG programs normally draw ticks floating in space.
This option connects the ticks with a solid axis.
When several adjacent points occur on a diagonal,
QuickMatch speeds up the plot by connecting them with a line.
This option forces QuickMatch to avoid this shortcut and plot all of the dots.
When you are checking for perfect matches it is not necessary to examine the sequence alignment.
This options turns of the alignment,
and just reports the existence of a perfect overlap on a diagonal.
Excludes any hits with the same name as the current search sequence.
This allows QuickMatch to be used to find overlaps in a private (Quick indexed)
database very easily.
Only reports the hits that give a perfect match.
This is a very fast option because QuickMatch first determines the best diagonal of comparison using a WordSearch
algorithm.
This option simply checks this one diagonal for a perfect match.
There is no sequence length limit in this case.
You can also produce sequence alignments as with QuickShow but limit the output to those above a specified stringency.
This checks the "Ratio" (defined as Quality/Length)
of the alignment and does not display it if it is below the specified stringency.
The next three options let you set the standard parameters for alignments:
Alignments
normally allow as many gaps as the GCG Procedure Library alignment routines can handle.
The gaps can be limited to gaps that shift the sequences out of phase from one another by a maximum of 10 base pairs from the diagonal that has the most points on it.
This option lets you expand the gap shift limits up to the length of the sequences being aligned.
If you specify a limit that is too high for any pair of sequences,
that pair will use the maximum possible gap limit instead.
sets the penalty for the creation of a gap.
sets the penalty per base pair for the extension of a gap.
The next four options affect the format of the optimal alignments:
The paired output file from this program displays sequence similarity by printing one of three characters between similar sequence symbols: a pipe character(|),
a colon (:),
or a period (.).
Normally a pipe character is put between symbols that are the same,
a colon is put between symbols whose comparison value is greater than or equal to 0.50,
and a period is put between symbols whose comparison value is greater than or equal to 0.10.
You can change these match display thresholds from the command line.
The three parameters for -PAIr
are the display thresholds for the pipe character,
colon,
and period.
The match display criterion for a pipe character changes from symbolic identity (the default)
to the quantitative threshold you have set in the first parameter.
A pipe character will no longer be inserted between identical symbols unless their comparison values are greater than or equal to this threshold.
If you still want a pipe character to connect identical symbols,
use x instead of a number as the first parameter.
(See the Data Files manual for more information about scoring matrices.)
When you print the output from this program,
it may cross from one page to another in a frustrating way -- especially when you print on individual sheets.
This option adds form feeds to the output file in order to try to keep clusters of related information together.
You can set the number of lines per page by supplying a number after the -PAGe
qualifier.
puts 50 sequence symbols on each line of the output file.
You can set the width to anything from 10 to 150 symbols.
suppresses large gap abbreviations,
showing all the sequence characters across from large gaps.
Usually,
gaps that extend one sequence by more than one complete line of output are abbreviated with three dots arranged in a vertical line.
This program normally monitors its progress on your screen.
However,
when you use the -Default option to suppress all program interaction,
you also suppress the monitor.
You can turn it back on with this option.
If your program is running in batch,
the monitor will appear in the log file.
If the monitor is slowing the program down,
suppress it with -NOMONitor.
writes a summary of the program's work to the screen when you've used the -Default qualifier to suppress all program interaction.
A summary typically displays at the end of a program run interactively.
You can suppress the summary for a program run interactively with -NOSUMmary.
Use this qualifier also to include a summary of the program's work in the log file for a program run in batch.
sets the number of bases or amino acids per 100 platen units (PU).
This is usually equivalent to the number of bases or amino acids per page.
Output from different GCG graphics programs that are run at the same density can be compared by lining up the plots on a light box.
These options apply to all GCG graphics programs.
These and many others are described in detail in Chapter 5,
Using Graphics of the User's Guide.
writes the plot as a text file of plotting instructions suitable for input to the Figure
program instead of drawing the plot on your plotter.
draws all text characters on the plot using Font 3 (see Appendix I)
.
draws the entire plot with the pen in stall 1.
These options let you expand or reduce the plot (zoom),
move it in either direction (pan),
or rotate it 90 degrees (rotate).
expands the plot by 20 percent by resetting the scaling factor (normally 1.0)
to 1.2 (zoom in).
You can expand the axes independently with -XSCAle and -YSCAle.
Numbers less than 1.0 contract the plot (zoom out).
moves the plot to the right by 30 platen units (pan right).
moves the plot up by 30 platen units (pan up).
rotates the plot 90 degrees.
Usually,
plots are displayed with the horizontal axis longer than the vertical (landscape).
Note that plots are reduced or enlarged,
depending on the platen size,
to fill the page.
Printed: April 22,
1996 15:55 (1162)
COMMAND-LINE SUMMARY
Minimum Syntax: % quickmatch [-INFile=]ggammacod.quick -Default
Prompted Parameters:
-MENu=2 1 is for dot-plots, 2 for optimal alignments
3 for list of hits
[-OUTfile=]ggammacod.match (output file for alignments only)
Local Data Files:
[-DATa=]quickdna.cmp the symbol comparison table for alignments
Command line options:
-MONitor display progress to terminal
-SUMmary display summary statistics at end
-PLOt dotplot only
-WORd=8 sets the match-criterion for a point
-RANdom plots all points, not just those on long diagonals
-CAPtion adds a caption to the plot
-NOSHOWalign show hits but no alignment
-NOSELF ignore hits with identical names
-PERFect show only perfect matches
-STRingency=1.0 minimum quality ratio to display
-WHOle show full alignment (default is bestfit alignment)
-LIMit=10 limit shift to +/- 10 in alignment
All GCG graphics programs accept these and other switches. See the Using
Graphics chapter of the USERS GUIDE for descriptions.
-FIGure[=FileName] stores plot in a file for later input to FIGURE
-FONT=3 draws all text on the plot using font 3
-COLor=1 draws entire plot with pen in stall 1
-SCAle=1.2 enlarges the plot by 20 percent (zoom in)
-XPAN=10.0 moves plot to the right 10 platen units (pan right)
-YPAN=10.0 moves plot up 10 platen units (pan up)
-PORtrait rotates plot 90 degrees
FUTURE DEVELOPMENT
ACKNOWLEDGEMENTS
LOCAL DATA FILES
OPTIONAL PARAMETERS
-PLOt
-RANdom
-WORd=8
-CAPtion
-ALL
-TICKAXes
-DOTSonly
-NOSHOWalign
-NOSELF
-PERFect
-STRingency=1.0
-WHOle Performs a full Needleman and Wunsch alignment (like Gap) rather than
a Smith and Waterman alignment (like BestFit)
.
-LIMit=10
-GAPweight=5
-LENgthweight=0.1
-PAIr=1.0,0.5,0.1
-PAGe=64
-WIDth=50
-NOBIGGaps
-MONitor
-SUMmary
-DENsity=1000
-FIGure=programname.figure
-FONT=3
-COLor=1
-SCAle=1.2
-XPAN=30.0
-YPAN=30.0
-PORtrait