 |
Response of Escherichia coli growth rate to osmotic shock
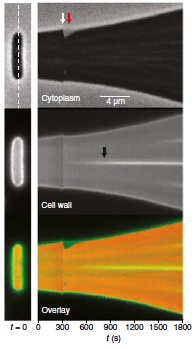
Abstract: It has long been proposed that turgor pressure plays an essential role during bacterial growth by driving mechanical expansion of the cell wall. This hypothesis is based on analogy to plant cells, for which this mechanism has been established, and on experiments in which the growth rate of bacterial cultures was observed to decrease as the osmolarity of the growth medium was increased. To distinguish the effect of turgor pressure from pressure-independent effects that osmolarity might have on cell growth, we monitored the elongation of single Escherichia coli cells while rapidly changing the osmolarity of their media. By plasmolyzing cells, we found that cell-wall elastic strain did not scale with growth rate, suggesting that pressure does not drive cell-wall expansion. Furthermore, in response to hyper- and hypoosmotic shock, E. coli cells resumed their preshock growth rate and relaxed to their steady-state rate after several minutes, demonstrating that osmolarity modulates growth rate slowly, independently of pressure. Oscillatory hyperosmotic shock revealed that although plasmolysis slowed cell elongation, the cells nevertheless “stored” growth such that once turgor was reestablished the cells elongated to the length that they would have attained had they never been plasmolyzed. Finally, MreB dynamics were unaffected by osmotic shock. These results reveal the simple nature of E. coli cell-wall expansion: that the rate of expansion is determined by the rate of peptidoglycan insertion and insertion is not directly dependent on turgor pressure, but that pressure does play a basic role whereby it enables full extension of recently inserted peptidoglycan.
|
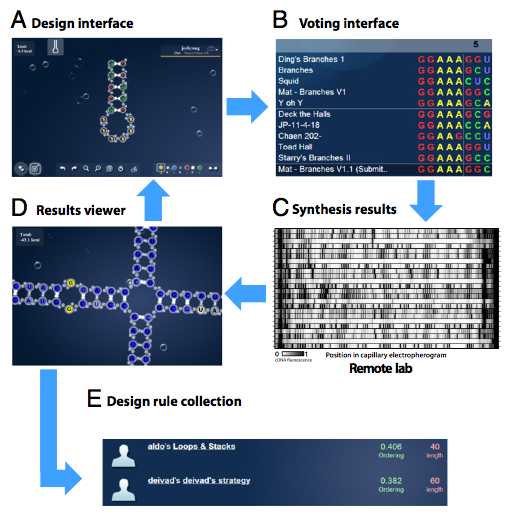 |
RNA design rules from a massive open laboratory
Abstract: Self-assembling RNA molecules present compelling substrates for the rational interrogation and control of living systems. However, imperfect in silico models—even at the secondary structure level—hinder the design of new RNAs that function properly when synthesized. Here, we present a unique and potentially general approach to such empirical problems: the Massive Open Laboratory. The EteRNA project connects 37,000 enthusiasts to RNA design puzzles through an online interface. Uniquely, EteRNA participants not only manipulate simulated molecules but also control a remote experimental pipeline for high-throughput RNA synthesis and structure mapping. We show herein that the EteRNA community leveraged dozens of cycles of continuous wet laboratory feedback to learn strategies for solving in vitro RNA design problems on which automated methods fail. The top strategies—including several previously unrecognized negative design rules—were distilled by machine learning into an algorithm, EteRNABot. Over a rigorous 1-y testing phase, both the EteRNA community and EteRNABot significantly outperformed prior algorithms in a dozen RNA secondary structure design tests, including the creation of dendrimer-like structures and scaffolds for small molecule sensors. These results show that an online community can carry out largescale experiments, hypothesis generation, and algorithm design to create practical advances in empirical science.
|
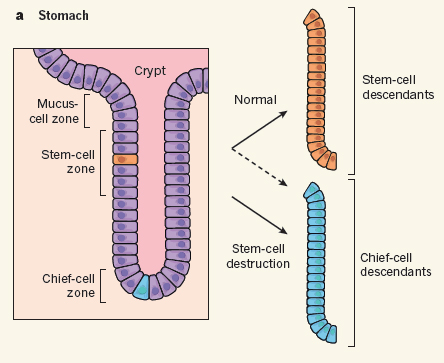 |
Differentiated cells in a back-up role
Abstract: When functional cells die, they are soon replaced. In most cases, the replacement cells arise either from the division of surviving mature cells of the same class or from the division and differentiation of tissue stem cells. But what happens when resident stem cells are selectively depleted? Two papers, one by Tata et al.1 published on Nature’s website today and the other by Stange et al.2 published in Cell, find that following depletion of stem cells in the stomach or lung, stem-cell function can be recovered through a surprising back-up function provided by specific differentiated cells in each tissue.
|
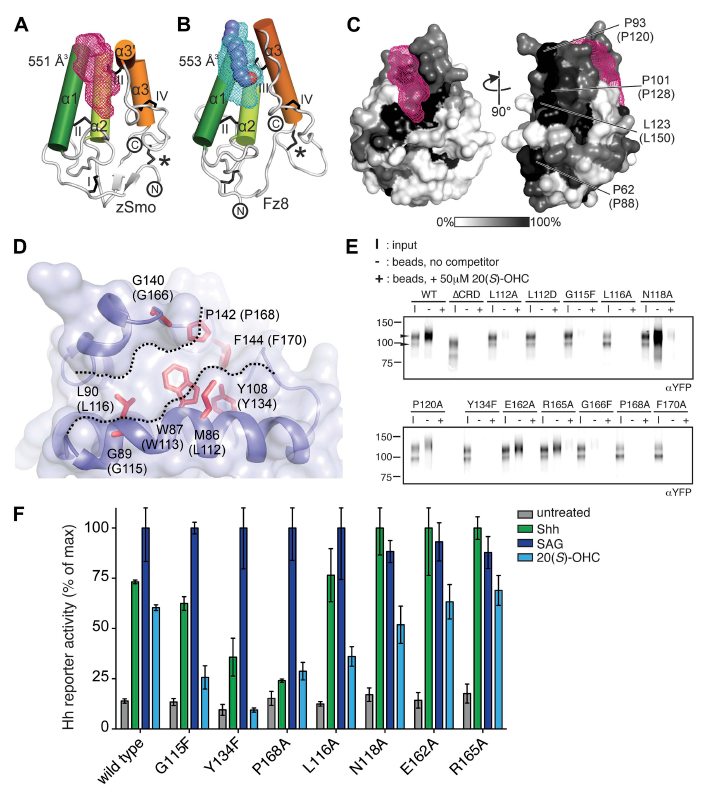 |
Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling
Abstract: The Hedgehog (Hh) signal is transduced across the membrane by the heptahelical protein Smoothened (Smo), a developmental regulator, oncoprotein and drug target in oncology. We present the 2.3 Å crystal structure of the extracellular cysteine rich domain (CRD) of vertebrate
Smo and show that it binds to oxysterols, endogenous lipids that activate Hh signaling. The oxysterol-binding groove in the Smo CRD is analogous to that used by Frizzled 8 to bind to the palmitoleyl group of Wnt ligands and to similar pockets used by other Frizzled-like CRDs to bind hydrophobic ligands. The CRD is required for signaling in response to native Hh ligands, showing
|
 |
TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends
Abstract: Telomere synthesis in cancer cells and stem cells involves trafficking of telomerase to Cajal bodies, and telomerase is thought to be recruited to telomeres through interactions with telomere-binding proteins. Here, we show that the OB-fold domain of the telomere-binding protein TPP1 recruits telomerase to telomeres through an association with the telomerase reverse transcriptase TERT. When tethered away from telomeres and other telomerebinding proteins, the TPP1 OB-fold domain is sufficient to recruit telomerase to a heterologous chromatin locus. Expression of a minimal TPP1 OBfold inhibits telomere maintenance by blocking access of telomerase to its cognate binding site at telomeres. We identify amino acids required for the TPP1-telomerase interaction, including specific loop residues within the TPP1 OB-fold domain and individual residues within TERT, some of which are
|
 |
Structural ensemble and microscopic elasticity of freely diffusing DNA by direct measurement of fluctuations
Abstract: Precisely measuring the ensemble of conformers that a macromolecule populates in solution is highly challenging. Thus, it has been difficult to confirm or falsify the predictions of nanometer-scale dynamical modeling. Here, we apply an X-ray interferometry technique to probe the solution structure and fluctuations of B-form DNA on a length scale comparable to a protein-binding site. We determine an extensive set of intrahelix distance distributions between pairs of probes placed at distinct points on the surface of the DNA duplex. The distributions of measured distances reveal the nature and extent of the thermally driven mechanical deformations of the helix. We describe these deformations in terms of elastic constants, as is common for DNA and other polymers. The average solution structure and microscopic elasticity measured by X-ray interferometry are in striking agreement with values derived from DNA–protein crystal structures and measured by force spectroscopy, with one exception. The observed microscopic torsional rigidity of DNA is much lower than is measured by single-molecule twisting experiments, suggesting that torsional rigidity increases when DNA is stretched. Looking forward, molecular-level interferometry can provide a general tool for characterizing solution-phase structural ensembles.
|
|
Genomic responses in mouse models poorly mimic human inflammatory diseases.
Abstract: A cornerstone of modern biomedical research is the use of mouse models to explore basic pathophysiological mechanisms, evaluate new therapeutic approaches, and make go or no-go decisions to carry new drug candidates forward into clinical trials. Systematic studies evaluating how well murine models mimic human inflammatory diseases are nonexistent. Here, we show that, although acute inflammatory stresses from different etiologies result in highly similar genomic responses in humans, the responses in corresponding mouse models correlate poorly with the human conditions and also, one another. Among genes changed significantly in humans, the murine orthologs are close to random in matching their human counterparts (e.g., R2 between 0.0 and 0.1). In addition to improvements in the current animal model systems, our study supports higher priority for translational medical research to focus on the more complex human conditions rather than relying on mouse models to study human inflammatory diseases. PRESS COVERAGE
|
|
Radial Construction of an Arterial Wall Abstract: Some of the most serious diseases involve altered size and structure of the arterial wall. Elucidating how arterial walls are built could aid understanding of these diseases, but little is known about how concentric layers of muscle cells and the outer adventitial layer are assembled and patterned around endothelial tubes. Using histochemical, clonal, and genetic analysis in mice, here we show that the pulmonary artery wall is constructed radially, from the inside out, by two separate but coordinated processes. One is sequential induction of successive cell layers from surrounding mesenchyme. The other is controlled invasion of outer layers by inner layer cells through developmentally regulated cell reorientation and radial migration. We propose that a radial signal gradient controls these processes and provide evidence that PDGF-B and at least one other signal contribute. Modulation of such radial signaling pathways may underlie vessel-specific differences and pathological changes in arterial wall size and structure.
|
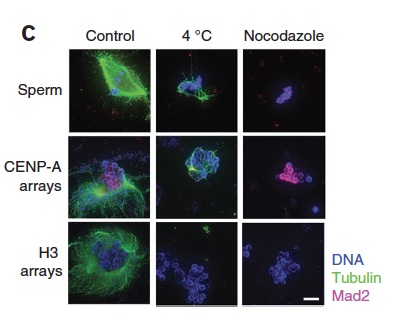 |
A cell-free system for functional centromere and kinetochore assembly Abstract: This protocol describes a cell-free system for studying vertebrate centromere and kinetochore formation. We reconstitute tandem arrays of centromere protein A (CENP-A) nucleosomes as a substrate for centromere and kinetochore assembly. These chromatin substrates are immobilized on magnetic beads and then incubated in Xenopus egg extracts that provide a source for centromere and kinetochore proteins and that can be cycled between mitotic and interphase cell cycle states. This cell-free system lends itself to use in protein immunodepletion, complementation and drug inhibition as a tool to perturb centromere and kinetochore assembly, cytoskeletal dynamics, DNA modification and protein post-translational modification. This system provides a distinct advantage over cell-based investigations in which perturbing centromere and kinetochore function often results in lethality. After incubation in egg extract, reconstituted CENP-A chromatin specifically assembles centromere and kinetochore proteins, which locally stabilize microtubules and, on microtubule depolymerization with nocodazole, activate the mitotic checkpoint.
|
|
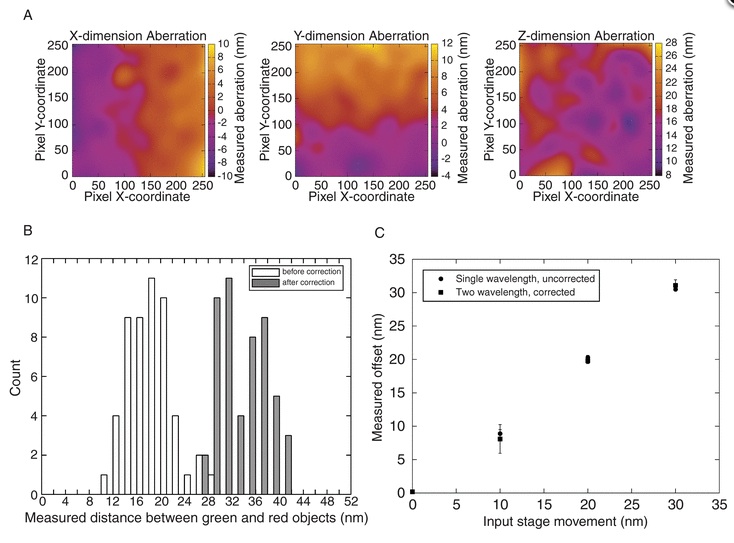
Imaging nanometre-scale structure in cells using in situ aberration correction
Abstract: Accurate distance measurements of cellular structures on a length scale relevant to single macromolecules or macromolecular complexes present a major challenge for biological microscopy. In addition to the inherent challenges of overcoming the limits imposed by the diffraction of light, cells themselves are a complex and poorly understood optical environment. We present an extension of the high-resolution colocalization method to measure three dimensional distances between diffraction-limited objects using standard widefield fluorescence microscopy. We use this method to demonstrate that in three dimensions, cells intrinsically introduce a large and variable amount of chromatic aberration into optical measurements. We present a means of correcting this aberration in situ [termed ‘Colocalization and In-situ Correction of Aberration for Distance Analysis’ (CICADA)] by exploiting the fact that there is a linear relationship between the degree of aberration between different wavelengths. By labelling a cellular structure with redundantly multi-colour labelled antibodies, we can create an intracellular fiducial marker for correcting the individual aberrations between two different wavelengths in the same cells. Our observations demonstrate that with suitable corrections, nanometre scale three-dimensional distance measurements can be used to probe the substructure of macromolecular complexes within cells. |
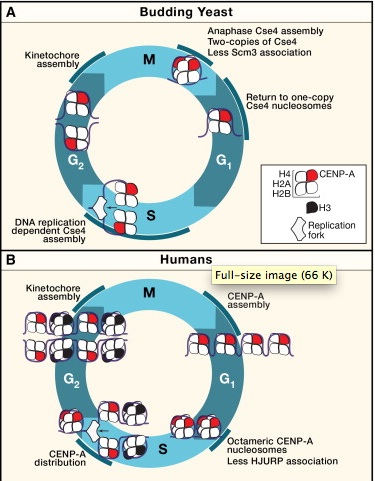 |
The split personality of CENP-A nucleosomes
A specialized chromatin domain, called the centromere, ensures accurate chromosome segregation during mitosis. Centromeres are the foundation for the assembly of the kinetochore, the site on each chromosome that acts as the primary interface between the chromosomes and the microtubules of the mitotic spindle. Maintaining the centromere is therefore essential for chromosome stability. The chromatin mark that determines centromere identity is a specialized histone H3 variant called CENP-A (called Cse4 in the budding yeast Saccharomyces cerevisiae). Budding yeast assemble Cse4 into chromatin during each round of DNA replication, whereas vertebrate CENP-A nucleosome assembly is replication independent, occurring during telophase and G1. |
Publications Archive PubMed Site |
|