

THE THINGS WE SEND OUT
SOME OF OUR MOST REQUESTED VECTORS: NFAT SUBUNITS, Calcineurin and BAF (also called mSWI/SNF) SUBUNITS ARE AVAILABLE FROM ADDGENE AT http://www.addgene.org/Jerry_Crabtree
HNF-1a:.
An expression plasmid in pBJ5. (Courtois et al., 1987) (Baumhueter
et al., 1990) (Kuo et al., 1990) (Mendel and Crabtree, 1991)
HNF-1b: An expression plasmid
in pBJ5. (Courtois et al., 1987) (Baumhueter et al., 1990) (Kuo et al.,
1990) (Mendel and Crabtree, 1991) (Mendel et al., 1991)
DCoH:. Transcriptional cofactor for HNF-1 binds to the
amino terminus of HNF-1a and b and stablizes dimers allowing enhanced DNA
binding and resistance to degradation. (Mendel et al., 1991) This was one
of the first transcriptional cofactors to be cloned. The crystal structure
was done in collaboration with Tom Albers lab and shows a remarkable resemblence
to TBP. (Endrizzi et al., 1995)
Antibodies to HNF-1a and b are
no longer available.
The alternative splice regions of the g fibrinogen genes (Crabtree and Kant,
1981) (Crabtree and Kant, 1982) are no longer available but can be obtained from
the Davies Lab at the University of Washington.
The regulatory regions of the HNF-1a gene: (MODY3)
that are controlled by HNF-4 (MODY1), which Calvin Kuo
in the lab used to define the transcriptional cascade (Kuo et al., Nature
1992) that Graham Bell’s lab later showed was the cause of Maturity
Onset Diabetes of the Young (MODY) as shown in the illustration below.
A Transcriptional Hiearchy Involved in Hepatic Development and Diabetes

Human Protein C: cDNA clones and genomic clones (Plutzky
et al., 1986) are no longer available. However Jorge
Plutzky
may
still have them. Protein C is now being marketed
by Lilly as Xigeris for treatment of sepsis.
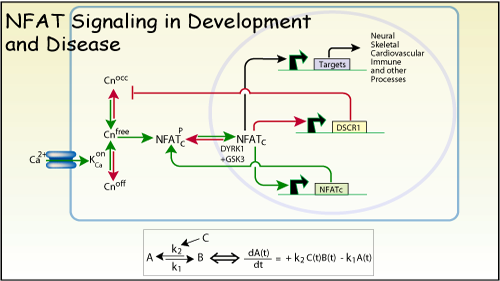
NFAT transcription complexes are assembled in the nucleus from cytoplasmic
(NFATc) and nuclear (NFATn) components (Flanagan et al Nature 1991) allowing
them to integrate signaling pathways (Crabtree, Science 1989). The cytoplasmic
subunits are dephosphorylated by calcineurin and are exquisitely cyclosporin-
and FK506-sensitive. A wide variety of signals result in Ca2+ increases,
activation of calcineurin and the dephosphorylation of the different NFATc
proteins. These signals include non-receptor tyrosine kinases such as the
T cell receptor, most tyrosine kinase receptors, non-concanonical wnt signals,
L type calcium channels, neurotropins, netrins and gap junctions. Indeed
it may be that this signaling pathway is the most widely used in vertebrates.
It has been difficult to detect genetically because of redundancy in the
components of the pathway and is also vertebrate-specific. Biochemically,
it is also difficult to detect in that most nuclear extract procedures
throw out the fractions that contain NFAT transcription complexes.(see
protocols) and a specialized extract procedure was originally developed
to detect it (Shaw et al Science 1988).IL-2 luciferase: Originally made
by David Durand and JP Shaw (Siebenlist et al., 1986). One plasmid has
the 650 bp fragment from Cla1 to a synthetic HindIII site at +51 in the
IL-2 gene. The other plasmid (15D Cx) has the same region truncated to
319 bp upstream of the cloned upstream of the luciferase gene (Durand et
al., 1987). Both behave the same in all of the assays we have used. The
mouse, human and Gibbon Ape regions (Durand et al., 1986) have all been
compared and show no difference in behavior. The clone 15D Cx has been
studied in transgenic mice and gives accurate expression (Emmel et al.,Science
1989)limited to activated T cells.
NF-AT luciferase: This plasmid has three copies of the NF-AT site cloned
upstream of the minimal IL-2 promoter from -89 to +51 (Shaw et al., 1988)
(Durand et al., 1988). This is the plasmid that was used to define NF-AT
and much of the Ca2+/calcineuruin/NF-AT signaling pathway (Emmel et al.,
1989) (Flanagan et al., 1991) (Clipstone and Crabtree, 1992) (Timmerman
et al., 1996)
NFIL-2A Luciferase. (Durand et al., 1988) (Ullman et al., 1991)Although
this plasmid binds the cJun protein and this site was used to purify JunD
(Ullman et al., 1993) it behaves very similar to the NF-AT luciferase plasmid
and NFAT transcription complexes with c-Jun may function at this site.
AP-1 Luciferase, Prepared from the sequences defined by Pam Mitchell
in the Bob Tjian’s laboratory from the SV-40 promoter. The actual
IL-2 AP-1 site is not very active in our hands.
NFkB Luciferase, Prepared from the kB enhancer as described from the Baltimore
laboratory. The IL-2 NFkB site is not active in our hands, however other
people have seen some activity with it.
CAML, Ca2+ modulatory Ligand of Cyclophilin. (Bram and Crabtree, 1994).
This plasmid can be obtained from Rick Bram at the University of Minnesota.
CalcineurinA Prepared by Neil Clipstone from the Murine clone encoding
the A1 subunit of calcineurin. (Clipstone and Crabtree, 1992). There are
three genes that encode the catalytic subunit.
Calcineurin B1 Prepared by Neil Clipstone from the murine
clone encoding the
B1 regulatory subunit of calcineurin. There are two genes that encode
the regulatory subunit. (Clipstone and Crabtree, 1992)
The NF-ATc Genes
The nomenclature is confusing for these proteins. The one below is the
original one (Flanagan et al Nature 1991 and Crabtree and Scbreiber 1992).
It is the offical nomenclature and can be found at the Genome Database
site: To find the new nomenclature look for: http://gdbwww.gdb.org/.
The cytoplamic, cyclosporin-senstive subunits are encoded by 4 genes.
The nuclear
subunits have not yet been purified but appear to be AP-1 and other proteins.
NF-ATc1(c), Expression vector for NF-ATc1 in a pBJ5 plasmid. The monoclonal
antibodies that we made to NF-ATc1 are sold by Pharmagen and Santa Cruz.
(Northrop et al., 1994). The crystal structure revealed that our initial
predictions that it was a rel-like protein were in fact correct. The
DNA binding domain is defective compared to a classic DNA binding domain
(Wolfe
et al., 1997) hence allowing the assembly of NFAT transcription complexes
to integrate signals.
NF-ATc2(p), Expression vector for human NF-ATc2 in a pBJ5 plasmid (Northrop
et al., Nature 1994). Sequence for the human and bovin clones were obtained
by Jeff Northrop and Steffan Ho.
NF-ATc3(x)(4), Expression vector for NF-ATc3 in a pBJ5
plasmid. (Ho et al., 1995). In this publication a full length protein
was cloned, but
it contains a start-site that is not present in most of the other isolates,
hence we thought it was not full lenght,but it actually is.
NF-ATc4(3), Expression vector for murine NF-ATc4 in a pBJ5 plasmid (Greaf
et al Nature 1999, Greaf et al Cell 2001 and Greaf et al Cell 2003).
TonEBP (NF-AT5)- Originally discovered by cloned by Kron and colleagues at John Hopkins. A cDNA was isolated by Steffan Ho, who now has these clones (see past lab members for address)
Prolyl Isomerases and Immunophilins
Cyclophilins A to D (Bram et al., 1993) These are expression vectors used by Rick Bram to define the cyclophilins that were active in blocking the actions of calcineurin.
FKBP12, 13, 25 and 52 were obtained from Stuart Schreiber’s laboratory
CRAC-Channel Deficient Cell Lines. These were isolated by Andrew Serafini
in the lab(Serafini et al., 1995). The mutations appear to be dominant
and have not yet been cloned. (Clipstone and Crabtree, 1992)
TAG Jurkats- This cell line was created by Steve Fiering and Jeff Northrop
in the lab and has SV-40 T antigen and a integrated NF-AT b gal plasmid
(Fiering et al., 1990), (Clipstone and Crabtree, 1992).
Yeast Genes Involved in the Rapamycin-Sensitive Growth Regulatory Pathway.
TOR1: (Zheng et al., 1995) (Fiorentino and Crabtree, 1997). Identical
to the clones isolated by M. Hall.
TOR2: (Zheng et al., 1995) (Fiorentino and Crabtree, 1997). Identical
to the clones isolated by M. Hall.
DNA-2 (ROT1) (Fiorentino and Crabtree, 1997)We have given our clones
and ts allels to Judith Campbell to distribute
Components of SWI/SNF-Like BAF Complexes
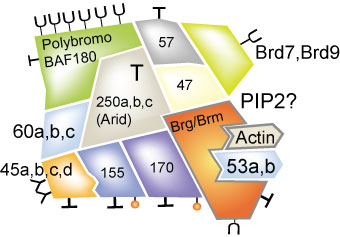
The names used are the ones that we originally used in characterizing the complex, however since neither SWI/SNF or BAF was acceptable to the Genome Nomenclature Committee we renamed them as "Swi-Related, Matrix-associated, Actin Dependent Regulators of Chromatin or SMARC protiens. To find the new nomenclature look for: http://gdbwww.gdb.org/
BAF250a, b,and c (SMARC d1, d2, Arid2) These can be obtained from Weidong Wang now a senior investigator at the National Institutes of Health. BAF250c is also called Arid or BAF200.
BAF 190a (BRG1) (SMARCA4) (Khavari et al., 1993)
(Dunaief et al., 1994) (Zhao et al., 1998) This plasmid is an expression
vector for
the full
length BRG/BAF190 protein which is similar to Drosphila Brm and contains
the conserved ATPase domain in the yeast SWI2/SNF2 protein. We also have
a dominant negative (Khavari et al Nature 1993), which we distributed widely, but now know is a gain-of-function since it
gives a phenotype that is more severe than the knock out of both Brg
and Brm. Hence, we no longer recommend that the dominant negative be
used to analyze the function of this complex.
HBrm- BAF190b (SMARCA2) This clone was isolated in Mose Yanif’s
lab at the Pasteur Insitute and is distributed by him. It is highly homologous
to Brg, but Brm complexes appear to have a different function than Brg
complexes based on the phenotypes of mice with mutations in these proteins.
Anti BRG1, JI is a rabbit antibody to the C terminus of BRG1. It is good
for most everything you need to do with this complex (Khavari et al.,
1993) (Dunaief et al., 1994) (Zhao et al., 1998).
BAF 170 (SMARCC2), This is an expression plasmid based on pBJ5. BAF170 has a
domain related to SWI3. (Wang et al., 1996)
BAF 155 (SMARCC1, This is an expression plasmid based on pBJ5.BAF155 has a domain
related to SWI3. (Wang et al., 1996)
BAF 60a, (SMARCD1) (Wang et al., 1996) This is an expression
plasmid based on pBJ5. BAF60a is related to a component of the yeast
SWI/SNF complex found by
Brad Crains and reported in Genes and Development 1996.
BAF 60b (SMARCD2)(Wang et al., 1996). This is an expression plasmid based on
pBJ5. BAF60a is related to a component of the yeast SWI/SNF complex
found by
Brad Crains and reported in Genes and Development 1996.
BAF 60c (SMARCD3)(Wang et al., 1996). This is an expression plasmid based on
pBJ5. BAF60a is related to a component of the yeast SWI/SNF complex
found by
Brad Crains and reported in Genes and Development 1996.
BAF57 (SMARCE1) (Wang et al., 1998). BAF57 has a kinesin coiled-coil domain
and a HMG domain. There is no homologue of this protein in the yeast
SWI/SNF
complex. BAF57 (SMARCE1) antibodies made in rabbits are also available.
These IP as well as Western blot and are also useful for immunofluorescene
and CHIP assays.
See Chi et al Nature 2002
BAF 53a (Actl6b) (Zhao et al., 1998). This is an actin related
member of of the ARP family of proteins. It directly interacts with BRG1
in the complex.
Shortly after we discovered this family member both Braid Crains and
Craig Peterson found yeast homologues in the yeast data base called ARP7
and ARP9 that are components of the yeast SWI complex. The yeast homologues
are less related to actin than BAF53a. Hence we do not yet know if there
is going to be a functional relationship between the yeast proteins and
the mammalian proteins.
BAF 53b (Actl6a) This subunit is neuron specific and has not been found to date
in other cell types. It is highly related to BAF53a.
BAF 47 (Kalpana et al., 1994) Also called INI for integrase interacting
protein or hSNF5 because it shares a domain with yeast SNF5 isolated
in Marion Carlson’s laboratory.
b-Actin. (Zhao et al., 1998). This is a component of the mammalian
complex but not the yeast complex. We feel that it is the most
distinctive feature
of the mammalian BAF complex and probably indicates that BAF is not
likely to be mechanistically related to the yeast complex. We have
b-actin with
mutations in the nuclear export sequence and also GFP tagged versions
of the protein.
Brd7 A newly discovered subunit not yet found in yeast or flies.
Brd9 A newly discovered subunit essential for the activity of the complex in es cells.
