
| ALL PUBLICATIONS | |
|
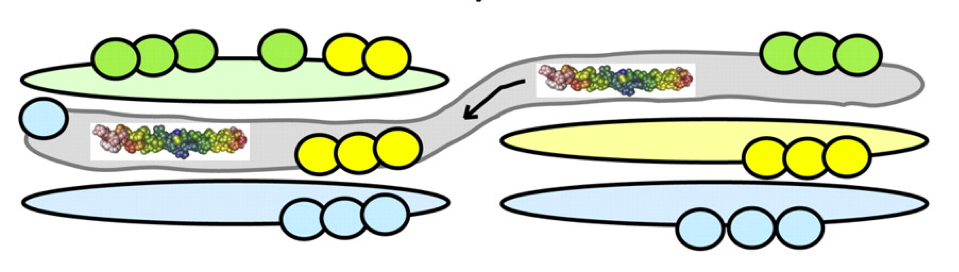
How the Golgi works: A cisternal progenitor model The Golgi complex is a central processing component in the secretory pathway of eukaryotic cells. This essential component processes more than 30% of the proteins encoded by the human genome, yet we will do not fully understand how the Golgi is assembled and how proteins pass through it. Recent advances in our understanding of the molecular basis for protein transport through the Golgi and within the endocytic pathway provide clues to how this complex organelle may function and how proteins may be transported through it. Described here is a possible model for transport of cargo through a tightly stacked Golgi that involves continual fusion and fission of stable, "like" subcompartments and provides a mechanism to grow the Golgi complex from a stable progenitor, in an ordered manner. READ FULL PUBLICATION
|
|
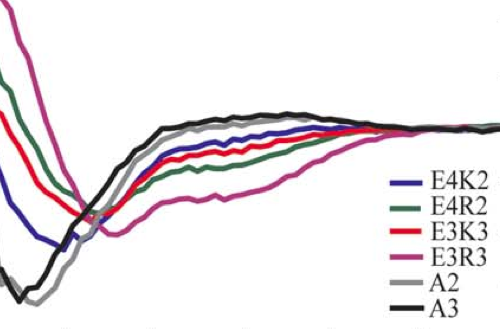
Helicity of short E-R/K peptides
|
 |
Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N Centromeres are specialized chromosonal domains that direct kinetochore assembly during mitosis. CENP-A (centromere protein A), a histone H3-variant present exclusively in centromeric nucleosomes, is thought to function as an epigenetic mark that specifies cetromere identity. Here we idently the essential centromere protein CENP-N as the first protein to selectively bind CENP-A nucleosomes but not H3 nuclesomes. CENP-N bound CENP-A-H4 tetramers. Mutations in CENP-N that reduced its affinity for CENP-A nucleosomes caused defects in CENP-N localization and had dominant effects on the recruitment of CENP-H, CENP-I and CENP-K to centromere. Depletion of CENP-N using siRHA (short interfering RNA) led to similar centromere assembly defects and resulted in reduced assembly of nascent CENP-A into centromeric chromatin. These data suggest that CENP-N interprets the information encoded within CENP-A nucleosomes and recruits other proteins to centromeric chromatin that are required for centromere function and propagation.
|